Goal of this exercise: Analyze the first study that used the CRISPR Cas9 system to successfully knock out an abnormal gene in human patients (Gillmore et al. 2021). Part 5 will determine the Cas9 cut location on the monkey TTR gene and look for possible frameshift mutations responsible for gene knockout in monkeys and humans.
Part 1: Overview of ATTR, CRISPR, and Gillmore et al. 2021
Part 2: Cut the human TTR gene with Cas9 under the direction of guide RNA
Part 3: Look for potential off-target effects of Cas9 in humans
Part 4: Examine mutations of patients associated with the study
Part 5: Analyze frameshift mutations in monkeys and humans
Overview of Part 5
Case It v7.0.4 will be used to determine Cas9 cut site and exon location on the monkey TTR gene and compare match percentages for monkey and human guide RNA target sites. The Genome Data Viewer will be used to look for frameshift mutations responsible for gene knockout.
Organization of Part 5
Step 1: Background information on non-human primate used in Gillmore’s study.
Steps 2-10: Determine Cas9 cut location on the monkey chromosome 18 sequence.
Steps 11-22: Use the Genome Data Viewer to identify targeted exon.
Clarification of how the Genome Data Viewer displays sequences.
Steps 23-24: Examine possible frameshift mutations responsible for gene knockout.
Steps 25-41: Compare matches for guide RNA target sequences on monkey and human TTR genes.
Concluding questions
Terminology
It may be helpful to watch this video before going through Part 5, as the terminology used for DNA transcription can be confusing.
1. Gillmore et al. 2021 tested their CRISPR/Cas9 system on crab-eating macaques (Macaca fascicularis), called ‘cynomolgus monkeys’ in their paper. Click here to learn more about this species.
Question: The use of NHP (non-human primates) in research has been controversial. This article proposes an ethical framework for such studies. What is your personal opinion regarding the ethics of such testing?
DETERMINE CAS9 CUT LOCATION ON MONKEY CHROMOSOME 18
The goal of the next series of steps (2 –10) is to determine the location of the guide RNA target on monkey chromosome 18, using the spacer sequence from Gillmore et al. 2021. You do not have to download this sequence as it has already been obtained from Genbank and is included with the Case It download.
2. If you have not already done so, download Case It v7.0.4 and follow the instructions on the PDF file to install it either on the Windows or Mac operating systems. Part 5 requires a version of Case It dated 10/03/21 or later. This can be determined by accessing the About menu at the top-left corner of the screen.
If Case It v7.0.4 is already open, quit the program and restart it. This is because you will soon be opening a very large file, so it is necessary to have a fresh start.
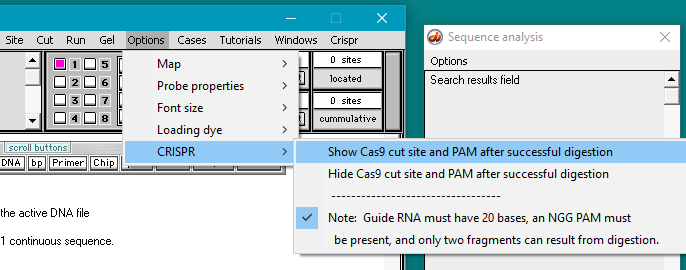
3. Click the Options menu and select CRISPR > Show Cas9 cut site and PAM after successful digestion. The default is to hide the Cas9 cut site, so if you quit the program and restart this option will have to be reactivated. If your version of Case It is dated earlier than 10/3/21, it will not have this feature (see Step 2), so if this is the case please re-download the program before continuing with this part of the exercise.
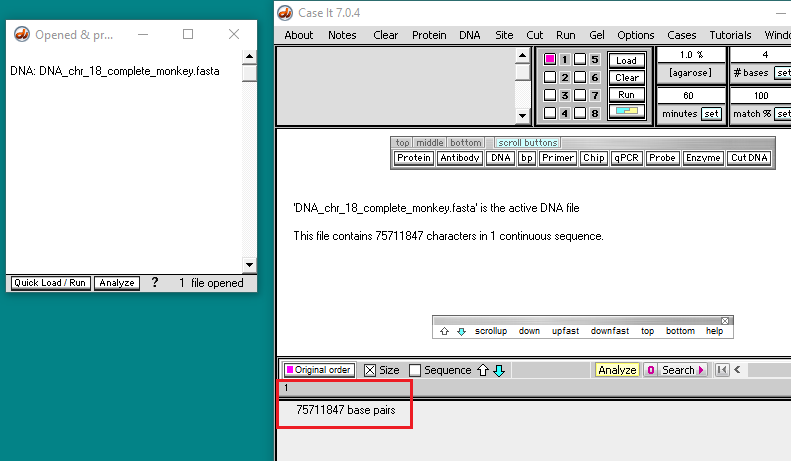
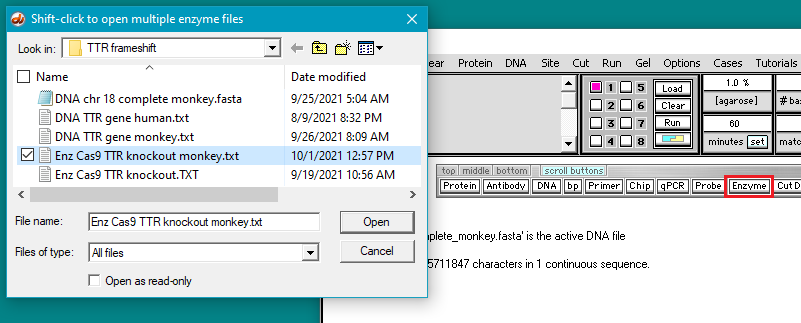
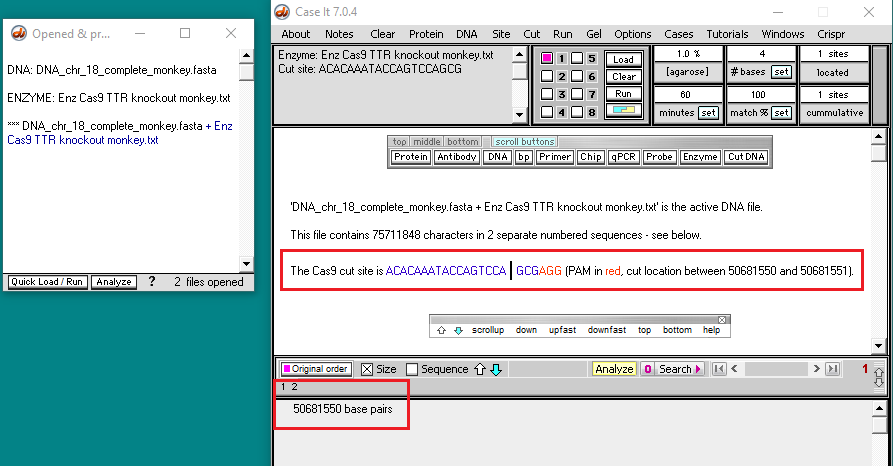
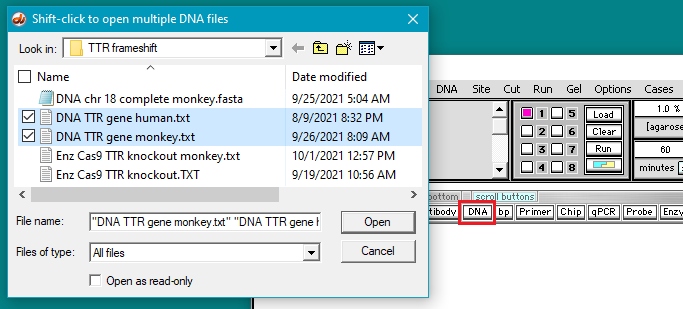
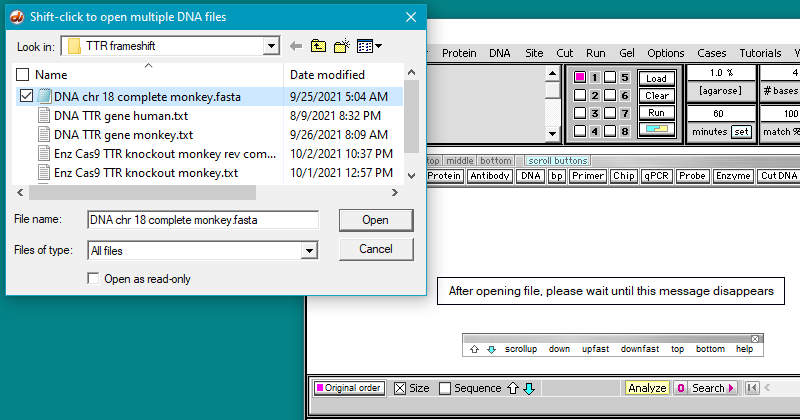
4. Click the DNA button on the silver button bar and select Case It v704 PC > TTR > TTR frameshift > DNA chr 18 complete monkey.fasta.
Because of the very large size of the file, it may take as long as 30 seconds to open the file. Wait for the “After opening file…” message to disappear before clicking anywhere on the program. This file is equivalent to a word processor document with over 18,000 pages.
5. The size of the opened file is 75711847 bp, representing the entire chromosome 18 sequence for Macaca fascicularis. Chromosome 18 is one of the smallest chromosomes in the monkey (and human) genome.
Questions: From Part 2, you know that the human chromosome 18 sequence has a size of 80,373,285 bp. What are some reasons for this apparent difference in size between human and monkey chromosomes?
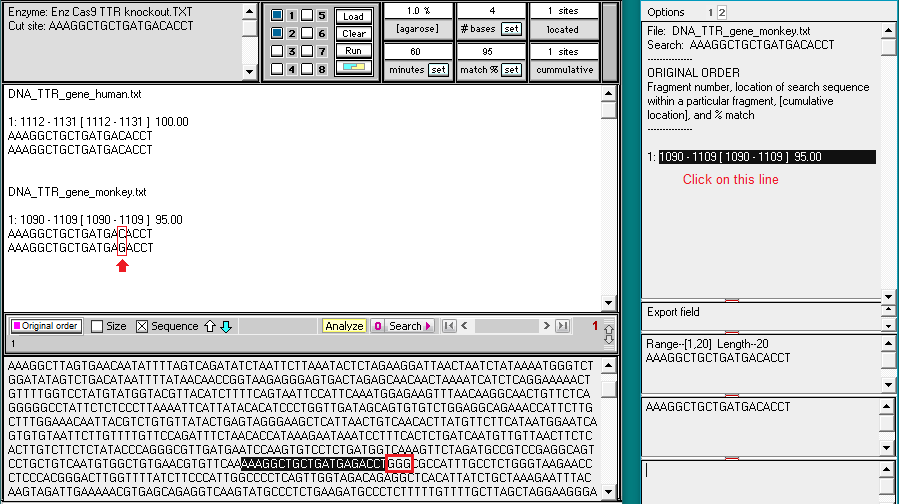
6. Click the Enzyme button and open the file Enz Cas9 TTR knockout monkey.txt.
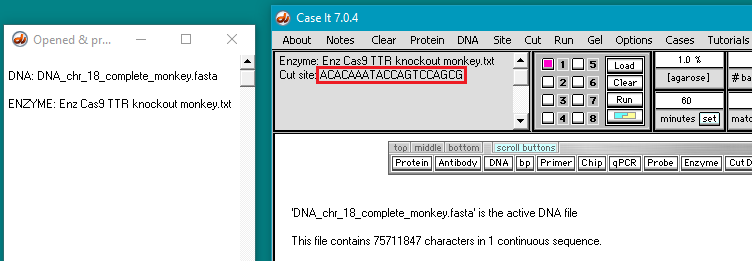
7. The enzyme file appears in the Opened & processed window, and the guide RNA target site appears in the gray field of the main screen. This target site for the monkey TTR gene was taken from Fig. 3B of Gillmore et al. 2012.
Note: A different target site was used for the human in vitro and in vivo trials -see Step 32 of Part 2.
Although files such as this are called ‘enzyme files’, they only consist of the guide RNA target site, as that is all that is required for Case It to find the location on a DNA sequence. In reality, Cas9 would have to be present along with the entire guide RNA molecule, as depicted in Part 1.
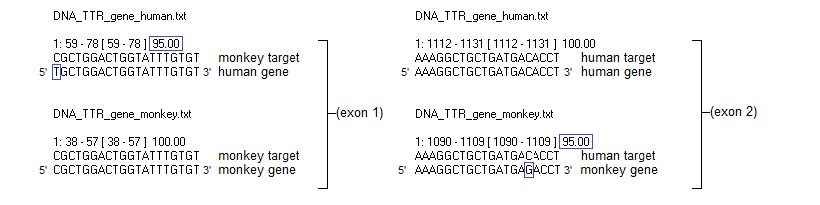
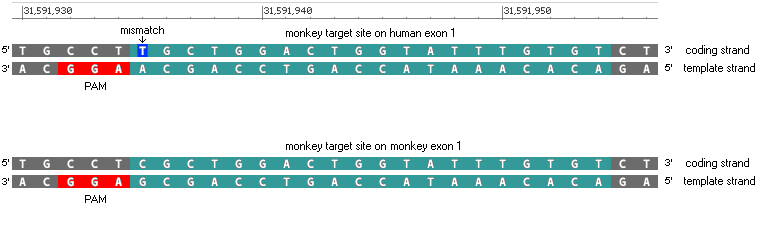
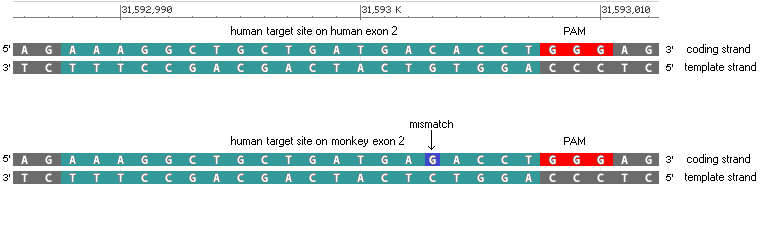
(After clicking these links, click the back arrow of your browser to return here.)
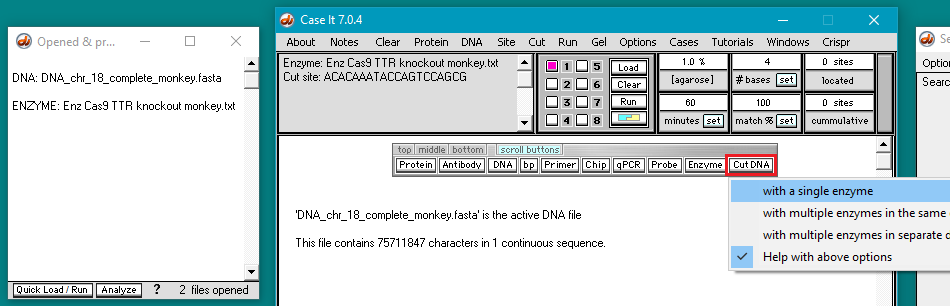
8. Click the Cut DNA button and select with a single enzyme.
Because of the very large size of the file, it will take about 10 seconds to digest the file. Wait for the “DNA digestion in process” message to disappear before clicking anywhere on the program.
9. Two fragments are produced by the digestion, indicating that only a single target site for the guide RNA was present on the entire chromosome 18 sequence. The size of the first fragment is 50681550, indicating that the Cas9 cut site is between 50681550 and 50681551.
For a cut to have occurred, an NGG PAM site must have been present next to the target site, as shown below (N stands for any of the four bases – A,T,C, or G). In this particular this case, the PAM is AGG. The cut location is designated by a vertical line.
Note: Case It v7.0.4 cannot display entire sequences when the file has more than 2,671,815 characters, but it can display the sequence for the cut site even for very large files (76 million characters in this case), as long as the option shown in Step 3 above is selected.
A gel will not be run with the two fragments because even though you can do it with the simulation, fragments this large would not separate on a real PFGE gel.
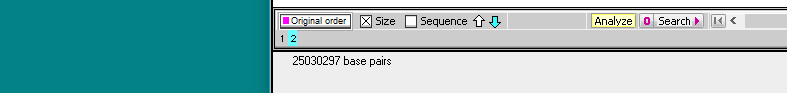
10. Click on the ‘2’ on the gray divider bar. The number that appears is 25030297, the size of the second fragment resulting from digestion. The sum of the two numbers is 75711847, the size of the uncut monkey chromosome 18 sequence (50681550 plus 25030297 equals 75711847 bp).
Note: The message in the white field in Step 9 states that the digested file contains 75711848 characters, not 75711847. This is because an extra return character was added by the program when the original sequence was digested.
USE THE GENOME DATA VIEWER TO IDENTIFY TARGETED EXON
The goal of the next series of steps (11-17) is to determine the exon targeted by the monkey guide RNA, using the Genome Data Viewer. This is different from the exon targeted for the human trials (see note under Step 7 above).
11. Open NCBI Entrez nucleotide (Genbank) by right-clicking on this link and selecting ‘open in new tab’ or ‘open in new window’.
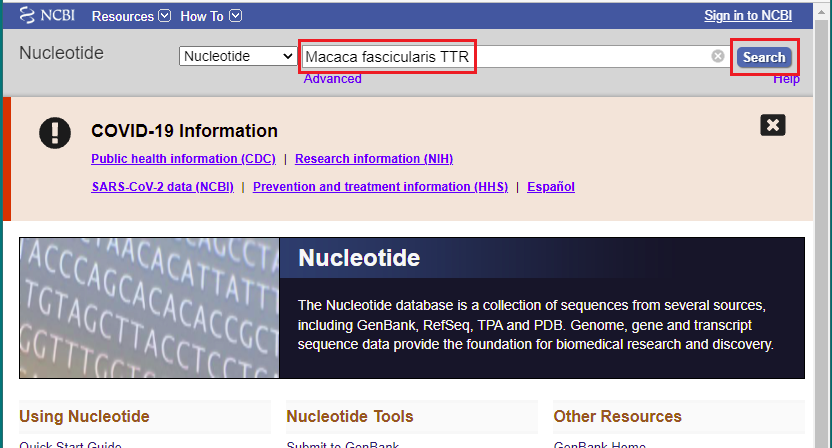
12. Copy and paste Macaca fascicularis TTR into the search field, then click the Search button.
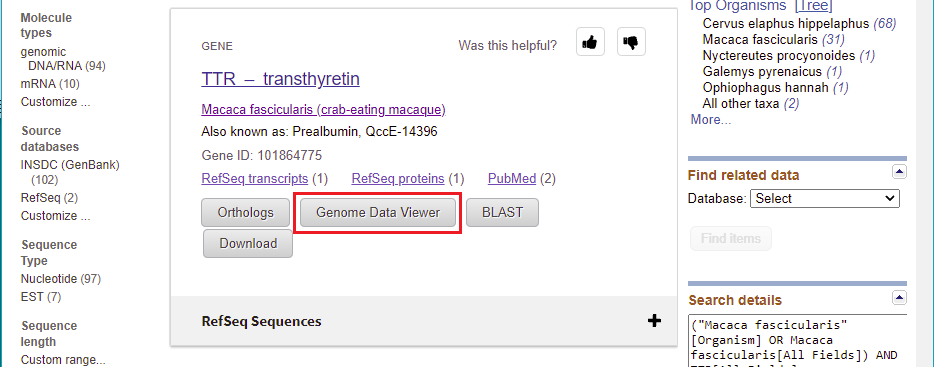
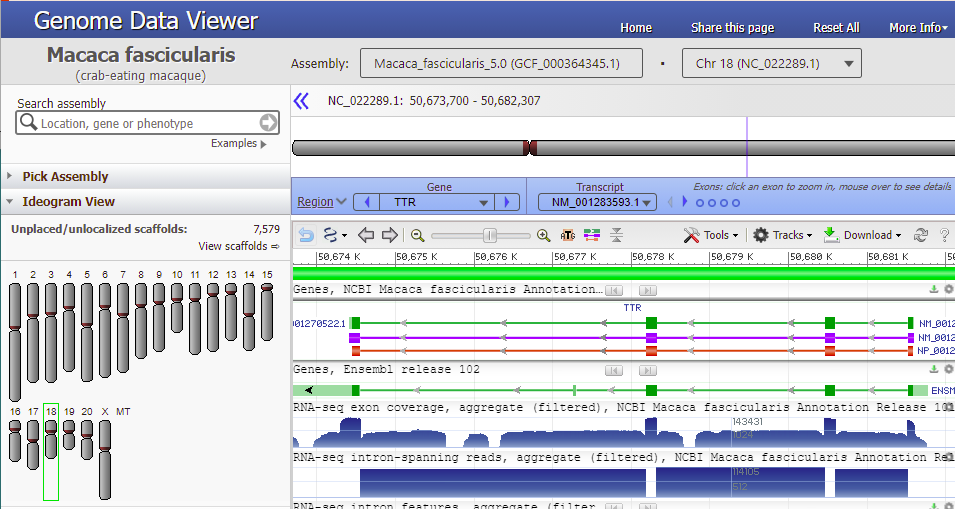
13. A window opens giving options for working with the monkey TTR gene. Click on the Genome Data Viewer button…
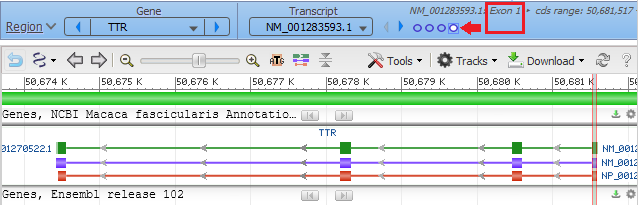
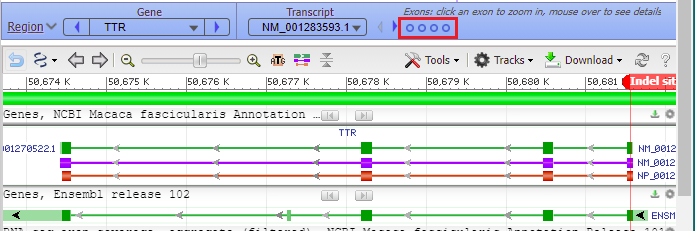
… and the viewer opens to the TTR gene of Macaca fasicicularis, the crab-eating macaque. Green boxes represent the four exons on the gene sequence, with narrow green lines representing introns between the exons. The purple line represents messenger RNA, and the orange line represent the protein precursor resulting from transcription and translation. The direction of the arrows indicates that the gene is being transcribed from right to left. This is the opposite of what you saw for the human TTR gene (Step 40-41 of Part 2), which had the arrows going from left to right*.
*Note: The exons on the monkey TTR gene appear to be in reverse order from the exons on the human TTR gene, but this is just an artifact of the way that the sequences were originally read into the Genbank database. Think of the DNA molecule as a ladder – either end of the ladder could be entered into the database, and opposite ends just happened to be read in for humans and monkeys. Humans and crab-eating macaques are closely related and share many of the same genes, with the same order of transcription.
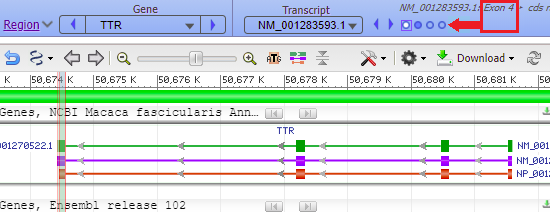
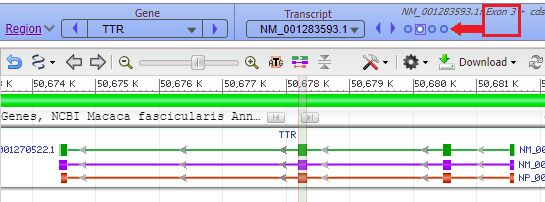
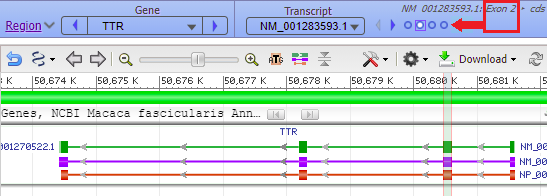
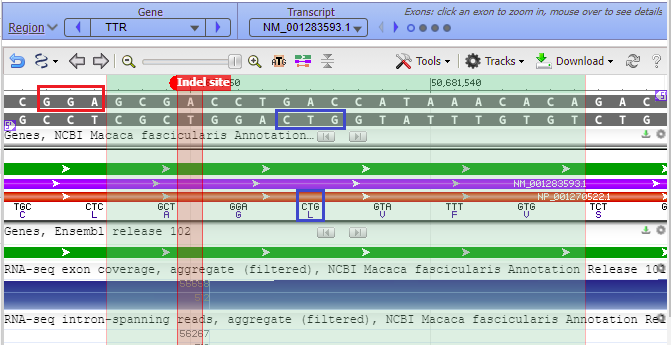
Hover your mouse cursor over the small circles in the blue bar. The message (in red box) changes from Exon 4 < Exon 3 < Exon 2 < Exon 1 and each exon is identified with a vertical bar, as shown in the series of screen shots below. Since the arrows on the lines point to the left, the exons are arranged from right to left, in the order of transcription and translation.
Note: There is a ‘flip strands’ feature that reverses the arrows so that they would point from left to right, in normal reading position. This feature will be shown later, but for now, leave the arrows pointing from right to left because that matches the way that Gillmore’s paper reported results of their analyses.
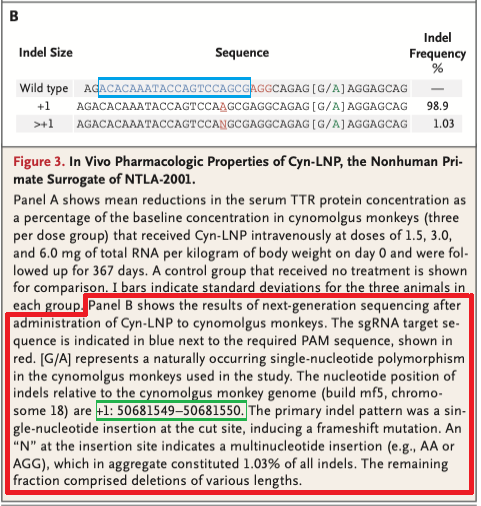
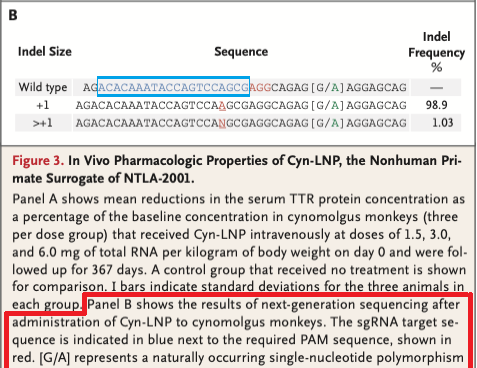
14. Before continuing, refer to Fig. 3B of Gillmore et al. 2021 (reproduced below). The indel location is given as 50681549-50681550* (green box), indicating that Cas9 cut between these locations, with the most common indel being a single base pair insertion (frequency 98.9%). Indels are insertions or deletions of nucleotides that occur when the cell attempts to repair the damage caused by Cas9 cutting the gene. Indels can cause frameshift mutations that disable the gene, as will be shown in Step 23 below.
*Note: Results from Step 9 above show that the chromosome 18 sequence was actually cut between locations 50681550-50681551, so that is the cut location that will be used in Step 16.
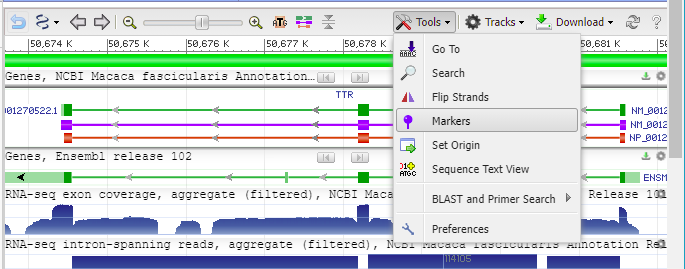
15. Click the Tools button and select Markers.
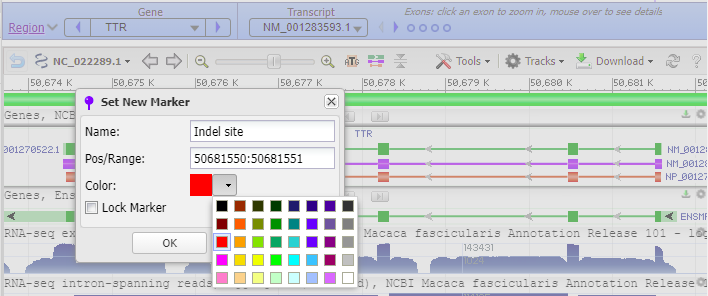
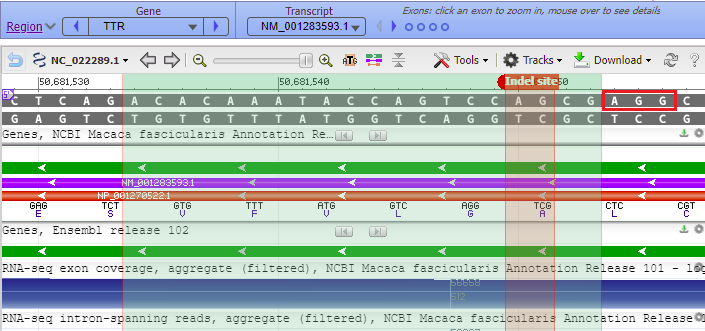
16. Copy and paste 50681550:50681551 into the Pos/Range box, type “Indel site” into the Name box, and select the color red for the indel marker. The Genome Data Viewer uses a colon (:) rather than a hyphen (-) to designate the range of a marker.
17. The marker appears at the far right, in exon 1 of the TTR gene (the order of exons was shown in Step 13).
Questions: Is this the same exon targeted by the guide RNA used for the human trials, as determined in Part 2? How might this affect interpretation of results of monkey vs. human trials?
(After clicking the link to Part 2, click the back arrow of your browser to return here.)
18. Refer again to Fig. 3B of Gillmore et al. 2021. The monkey guide RNA target sequence is shown in the first line below (labelled ‘Wild type’, in the blue box).
Copy the guide RNA target sequence: ACACAAATACCAGTCCAGCG to your computer’s clipboard.
Question: The guide RNA sequence target sequence used for the monkey trial (ACACAAATACCAGTCCAGCG) was different than the one used for the trial on human patients (AAAGGCTGCTGATGACACCT). Why did the investigators use different guide RNA sequences for the two trials?
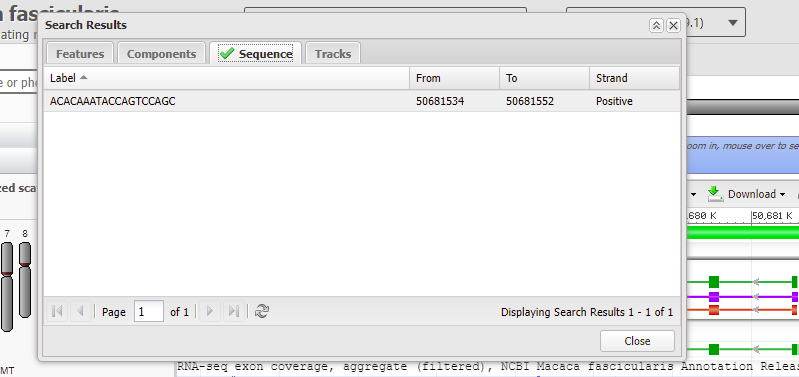
19. Click the Tools button again, but this time select Search.
20. Paste the copied guide RNA target sequence into the search field, then click OK.
21. After a few moments a green check mark will appear on the Sequence tab, indicating that the sequence was found. Click the Sequence tab, then double-click on the line showing the sequence, highlighted in gray below. Wait a few seconds, then click the Close box to hide the window.
22. The 20 bases of the guide RNA target site are highlighted in green, with the top strand matching the order in Fig. 3B (step 18), which was ACACAAATACCAGTCCAGCG. The PAM site (AGG in red box) is next to the green shaded target site. Recall from Step 9 that a PAM site must be present for Cas9 to cut. The Cas9 cut site would be three base pairs away from the PAM, located between the A and the G in the red-shaded Indel site marker.
Clarification of how the Genome Data Viewer displays sequences: Rules to keep in mind are explained in the blue section below, with a summary at the end of the section.
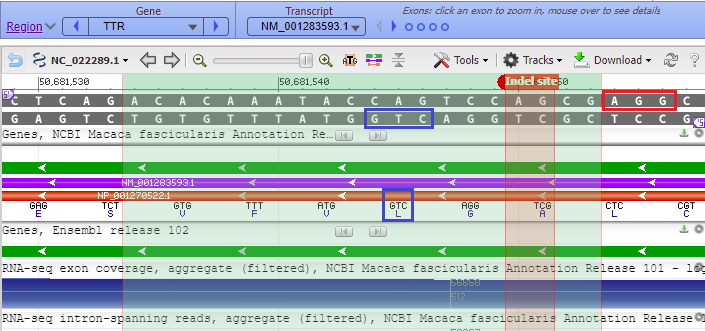
Arrows on the green, purple and orange bars represent the direction of transcription and translation of the DNA code, in the 5′ to 3′ direction. If the arrows point from right to left, the letters on the coding strand have to be read from right to left. The coding strand is the DNA strand that matches the three letters associated with each amino acid symbol (below the orange bar).
For example, the amino acid leucine (L) is associated with the DNA codon CTG, so the letters in the blue boxes need to be read right to left as CTG, not as GTC as first appears. That is because the 5′ end of the lower DNA strand is on the right, and DNA sequences are always read in the 5′ to 3′ direction. Small purple labels show the 5′ end of each strand. If the 5′ end is on the left side of the top strand, the 5′ end will be on the right side of the bottom strand, as shown in the example below.
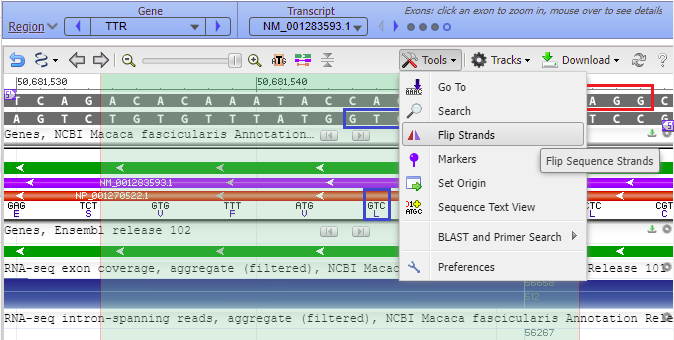
A command is available that flips the strands so that letters on the coding strand can be read from left to right. To activate this, click Tools and select Flip Strands…
The strands have been flipped, with the codon for leucine now read as you normally would, from left to right (CTG). The 5′ ends of each strand (small purple labels) have also been flipped, as is evident from a comparison of the image above with the image below. If the 5′ end is on the right side of the top strand, the 5′ end of the bottom strand will be on the left, as shown below. Again, transcription is always in the 5′ to 3′ direction. Think of the DNA molecule as a ladder that can be rotated (‘flipped’) horizontally – it is the same ladder, regardless of the way that you view it.
The guide RNA target site (shaded in green) and PAM (in red box) have been flipped, along with the orientation of the Indel site three bases away from the PAM. The PAM is still downstream (towards the 3′ end) of the guide RNA target site.
To summarize, here are the rules to remember:
(1) The DNA coding strand is the strand with letters that match the three letters associated with each amino acid (e.g. CTG associated with L, for Leucine – see image directly above).
(2) If the arrows point from right to left, the letters on the coding strand have to be read from right to left. If the arrows point from left to right, then the letters associated with the amino acid are read from left to right as you normally would. The arrows always point in the order of transcription (that is, the order in which DNA is transcribed into messenger RNA).
(3) You can use the ‘Flip Strands’ feature to reverse the strands so that the letters on the coding strand can be read from left to right.
—————————————————————
(4) The PAM can be on either the coding or non-coding (template) strand of the DNA molecule.
(5) Regardless of which strand it is on, the PAM sequence must always be downstream (towards the 3′ end) of the target sequence on that strand.
EXAMINE FRAMESHIFT MUTATIONS RESPONSIBLE FOR GENE KNOCKOUT
The goal of the next series of steps (23-24) is to determine if the CRISPR/Cas9 system can knock out (disable) the monkey TTR gene. Recall that the ‘Indel site’ marks the location where indels will occur when the cell repairs the cut site. The extra base(s) added can potentially cause a frameshift mutation that interferes with proper translation of the protein.
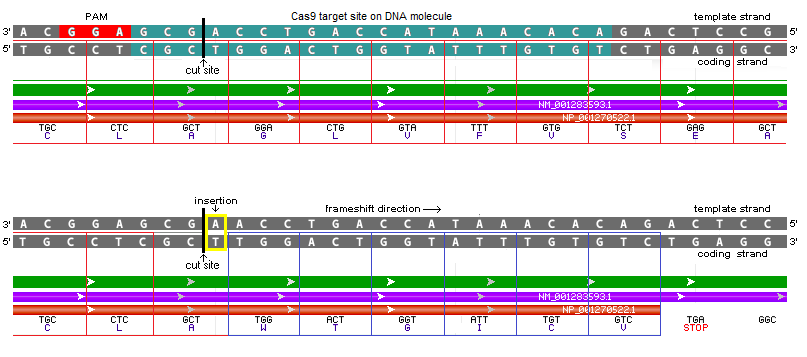
23. The most common indel for the monkey TTR cut site was a single base pair insertion (yellow box) at the location of the Cas9 cut site (black vertical line). This causes a frameshift resulting in the formation of a TGA stop codon (lower right). The stop codon stops translation of the amino acid sequence, thus ‘knocking out’ the gene. The image below shows the double-stranded DNA sequence before (top) and after (bottom) the single base pair is inserted.
Note: The diagram below was created by modifying an image from the Genome Data Viewer. The ‘Flip Strands’ feature was used to reverse the strands so that the letters on the coding strand could be read from left to right. The purple line represents the mRNA sequence, and the orange line is the protein precursor (amino acid sequence before modification into a functional protein).
Keep in mind that whether or not the DNA strands are flipped, it is the same DNA molecule. For example, in the image below (from Fig.3B of Gillman’s paper) the template strand is reversed compared to the image directly above, with the PAM (AGG in red letters) on the right in Fig.3B. Note: Red 5′ and 3′ labels were not part of the original figure, but were added to make it easier to compare Fig.3B with the frameshift diagram above. The PAM is always downstream (towards the 3′ end) of the strand on which it is located.
Question: The ‘Indel Frequency’ on the right of Fig. 3B shows that the vast majority (98.9%) of the indels were single base-pair insertions (A). 1.03% were insertions with more than one base, designated by the letter N. These insertions were either AA or AGG. Would either of these have resulted in a stop codon in exon 1 of the monkey TTR gene?
24. Search the human TTR gene for potential frameshift mutations. Here is a review of the procedure used above, modified for the human trial:
(1) Open the Entrez Nucleotide site.
(2) Search for Homo sapiens TTR.
(3) Click the Genome Data Viewer button.
(4) Use the Tools menu and search for the target site for the guide RNA used in the human trials: AAAGGCTGCTGATGACACCT
(5) Identify the Cas9 cut site, 3 bp upstream of the PAM site (GGG in this case), and look for potential stop codons downstream of the cut site.
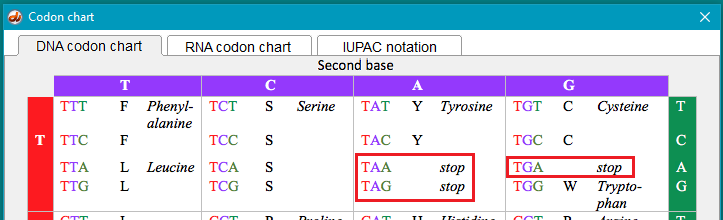
As a reference, click on the Search button to access a codon chart that includes stop codons.
Questions
(1) Assuming that the most common indel is a one bp insertion or deletion, can you find a stop codon in exon 2 of the human TTR gene? Remember that if one can be found, it must be downstream (toward the 3′ end) of the DNA sequence. The three DNA stop codons in humans are TGA, TAA, and TAG. Can you find a stop codon if the mutation had more than one insertion or deletion? Recall that in the monkey trials, 99% of the idels were a single base pair insertion.
(2) Is there a potential stop codon in exon 3? Considering the mechanism for splicing RNA after transcription but before translation, is it physically possible for a frameshift mutation in exon 3 to be induced by a frameshift mutation in exon 2?
(3) In the legend to Figure 2 of Gillmore et al. 2021, the authors say that the “primary indel patterns were a single-nucleotide deletion or insertion at the cut site, inducing a frameshift mutation (data not shown).” They are referring to human hepatocytes (liver cells) studied in vitro. Why did they not show the data for the human trials, when they did show the data for the monkey trials in Fig. 3B?
COMPARE GUIDE RNA TARGET SEQUENCES USED FOR MONKEYS AND HUMANS
The goal of steps 25-33 is to search the human and monkey TTR genes using the human guide RNA target sequence. The goal of steps 34-40 is to search the human and monkey TTR genes using the monkey guide RNA target sequence.
25. Quit and restart Case It v7.0.4 before continuing. Normally you would clear the program instead of restarting it, but anytime you have finished working with a very large file, such as a complete chromosome sequence, you should quit and restart the program.
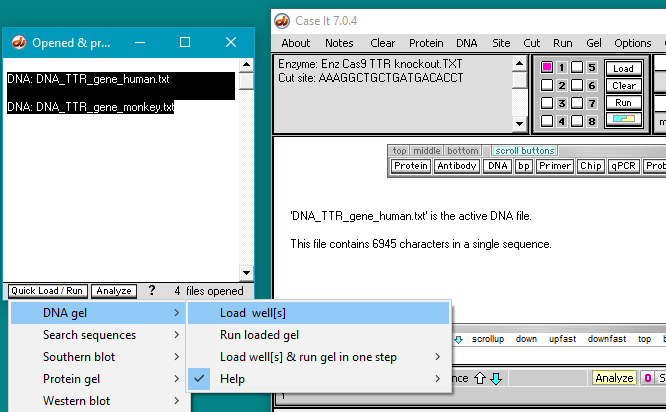
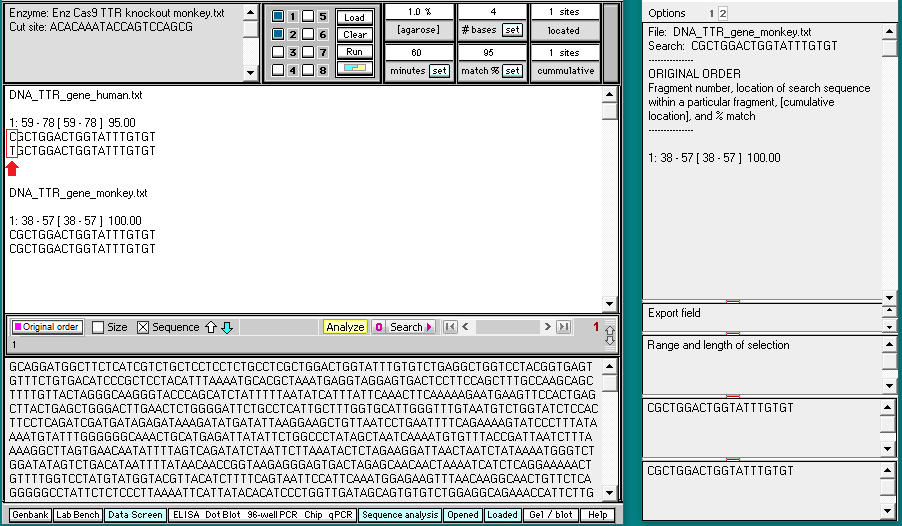
Click the DNA button on the silver button bar, then shift-click to select the files DNA_TTR_gene human.txt and DNA_TTR_gene_monkey.txt. They are located in the TTR frameshift folder of the TTR folder.
Double-click on the files or click the Open button.
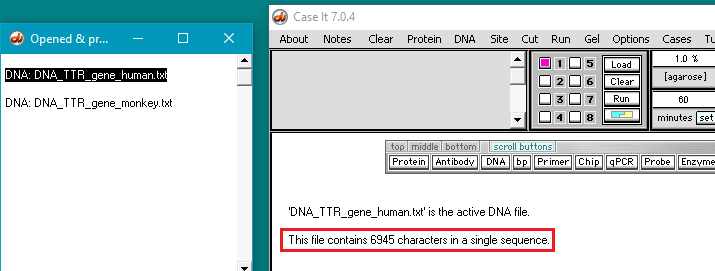
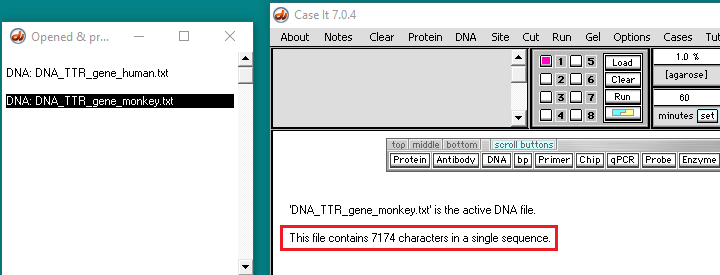
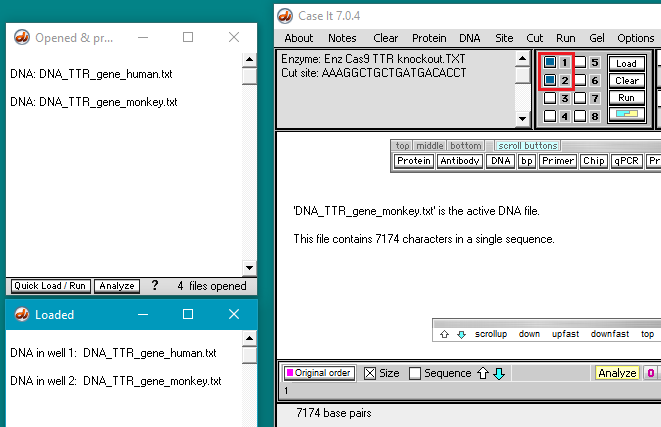
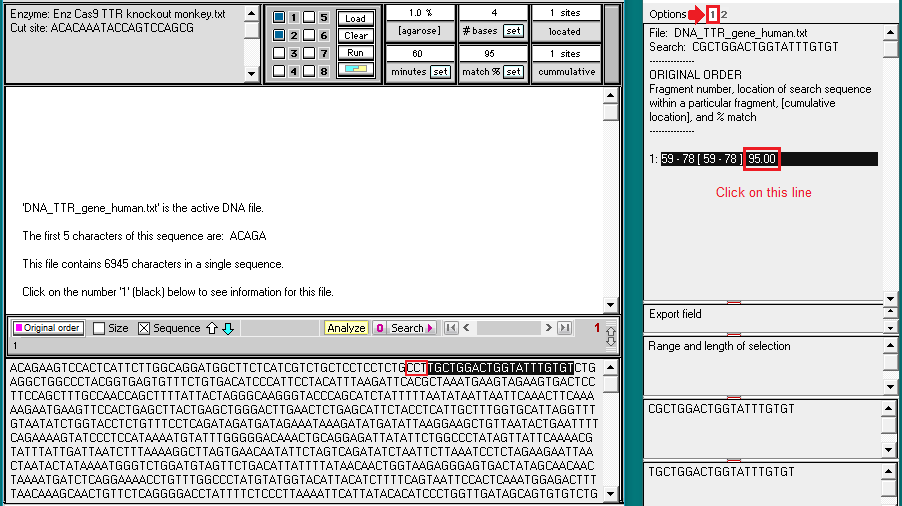
26. The filenames will appear as lines in the Opened & processed window. Click on the line ‘DNA: DNA_TTR_gene human.txt’ and verify that the human TTR gene has 6945 characters (each representing a base pair). Then click on the line ‘DNA: DNA_TTR_gene_monkey.txt‘ and verify that the monkey TTR gene has 7174 characters.
Question: What could account for the difference in size of the human and monkey TTR genes?
27. Shift-click to select both DNA files, then click the Quick Load / Run button and select DNA gel > Load well(s). The squares next to wells 1 and 2 turn blue indicating that they are loaded (second image below). and the contents of the loaded wells appear in the Loaded window.
Note: Wells were loaded because this a necessary step prior to searching DNA sequences. It is just the way that the search procedure works, and has nothing to do with running a gel.
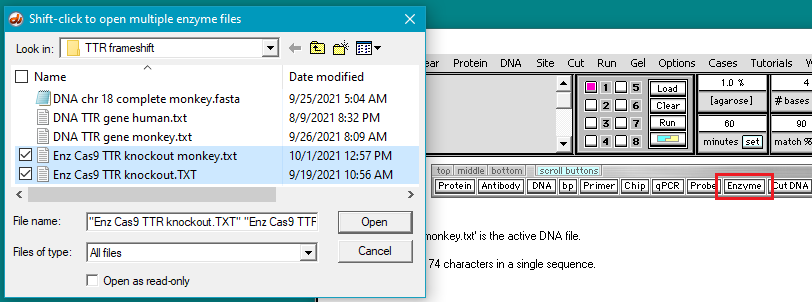
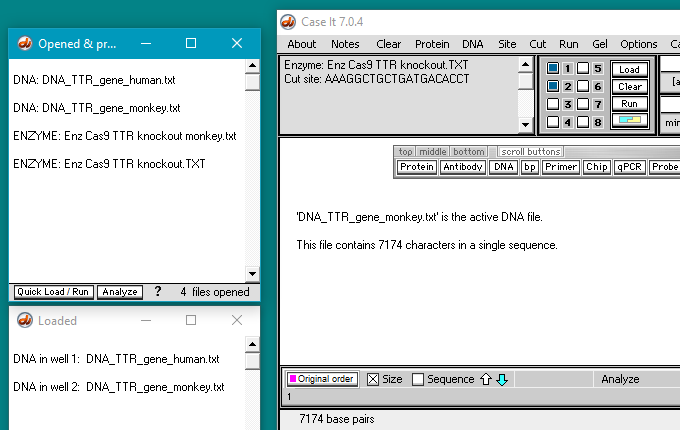
28. Click the Enzyme button and shift-click to open the files Enz Cas9 TTR knockout monkey.txt and Enz Cas9 TTR knockout.TXT. These two files contain the target guide RNA sequences for the monkey and human trials, respectively. The filenames for the enzyme files appear in the Opened & processed window (second image below).
29. Before continuing, the match percent needs to be lowered from 100% to 95%. Double-click (or drag) on the number 100 in the match % field to select it, then type in 95. Click the Set button and select Set match %. The reason for doing this will become clear later in the exercise.
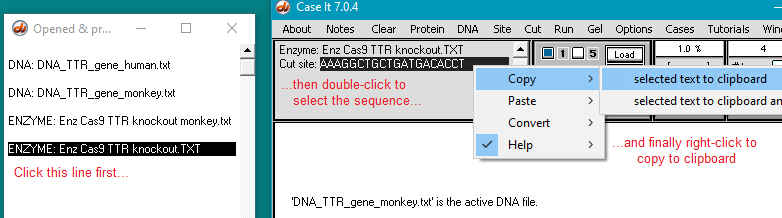
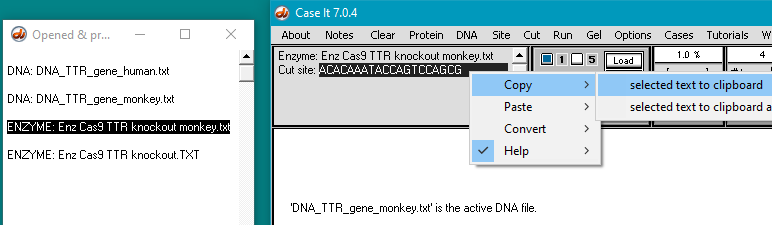
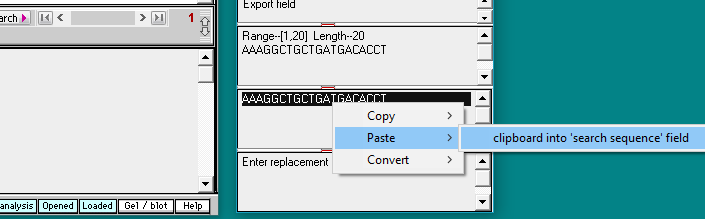
30. Use the following procedure to copy the human guide RNA target sequence to the clipboard:
(1) Click the line Enz Cas9 TTR knockout.TXT in the Opened & processed window. This contains the guide RNA target sequence for the human trials.
(2) Double-click on the sequence in the gray field to highlight it.
(3) Right-click on the sequence field to open the pop-up menu.
(4) Select Copy > selected text to clipboard.
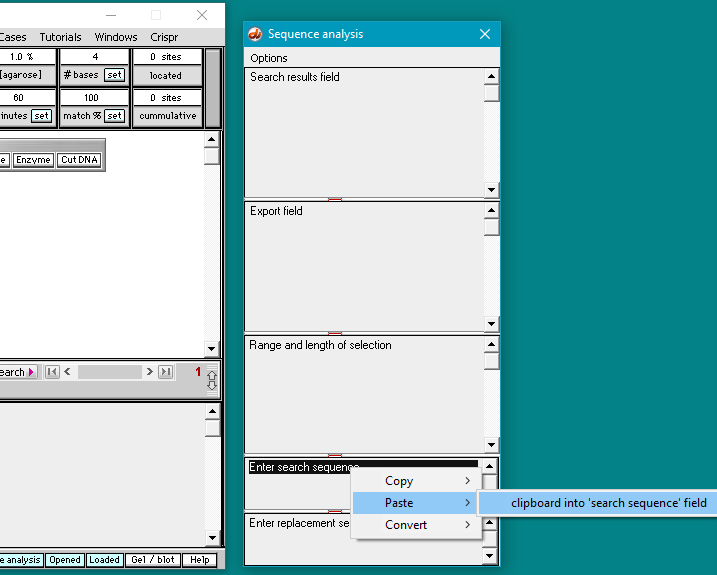
31. Use the following procedure to paste the human guide RNA target sequence into the Sequence analysis window:
(1) Right-click on the second field from the bottom in the Sequence Analysis window (this field is labelled ‘Enter search sequence’).
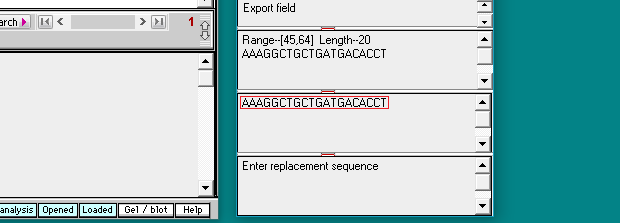
(2) Select Paste > clipboard into ‘search sequence’ field. The human guide RNA target sequence will appear in the field: AAAGGCTGCTGATGACACCT (second image below).
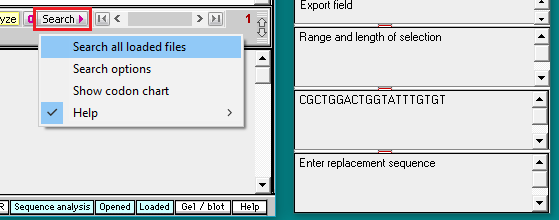
32. Click the Search button, select Search all loaded files, and wait a few seconds for the ‘Searching sequences’ message to disappear.
33. Click on the line in the Sequence analysis window (on right), and the location for this hit will be shown in at in the field at the bottom of the main screen. As expected, there is a 100% match (20/20) on the human TTR gene with the guide RNA sequence used in the human trials. There is a 95% match (19 out of 20) on the monkey TTR gene with the guide RNA sequence used in the human trials, the only difference being a C-G change at the position of the red arrow below. The match is on exon 2 of the monkey gene, as could be verified using the Genome Data Viewer. Although you could right-click on the screen to save this information (or take a screen shot), that will not be necessary as these results will be summarized below.
The goal of the next series of steps (34-40) is to search the human and monkey TTR genes using the monkey guide RNA target sequence.
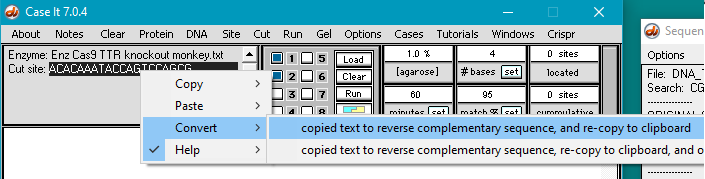
34. Use the following procedure to copy the monkey guide RNA target sequence to the clipboard:
(1) Click the line Enz Cas9 TTR knockout monkey.txt in the Opened & processed window. This contains the guide RNA target sequence for the monkey trials.
(2) Double-click on the sequence to select it.
(3) Right-click in the sequence field to open the pop-up menu.
(4) Select Copy > selected text to clipboard.
35. Convert the monkey guide RNA target site to its reverse complementary sequence by right-clicking on the sequence field and selecting Convert > copied text to reverse complementary sequence, and re-copy to clipboard as shown below. The reason for this is explained in Step 41 below.
36. Right-click on the search sequence field as you did in Step 31, then paste to replace the human target site with the monkey target site.
37. The monkey sequence appears as the reverse complement of the original sequence. Note that this matches the coding strand sequence in Step 23 above, also shown in the second image below. This will be explained in more detail in Step 41 below.
38. Click the Search button and select Search all loaded files.
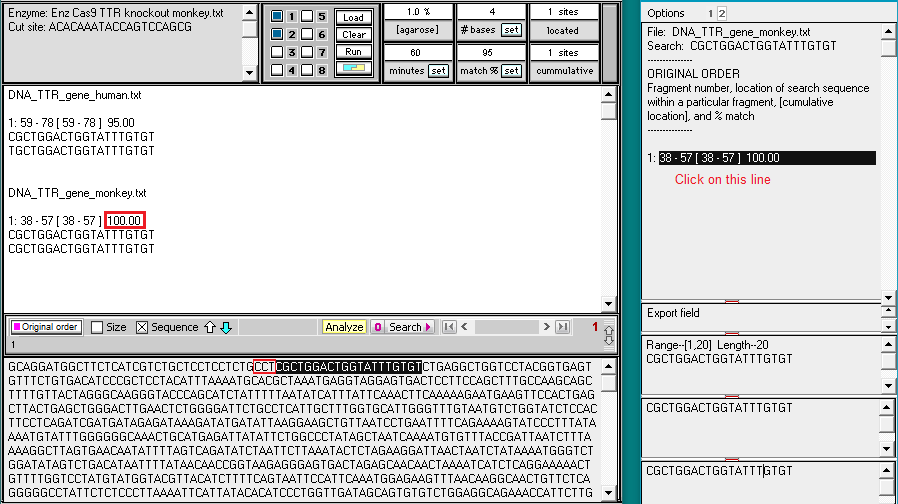
39. As you would expect, there is a 100% match between the monkey TTR gene and the monkey guide RNA target site. Click on the line as indicated below, and note the CCT (in red box) to the left of the highlighted target site at the bottom of the screen. These three bases are complementary to the PAM shown in Step 23 above (CCT on the coding strand is complementary to GGA on the template strand).
40. The summary on the main screen also indicates a 95% match between the human TTR gene and the monkey guide RNA target site, the only difference being a C-T change at the position of the red arrow. You do not need to save this result, as it is part of the summary in Step 41 below.
To see where the 95% match is located on the human gene, click the small number 1 at top right (in red box), then click the line as indicated below. There is a CCT sequence to the left of the target site, complementary to the GGA PAM site, just as there was in Step 39 for the monkey gene.
Note: Immediately after running a search, a summary of results appears as shown above. To then go back and forth between results for the human and monkey genes, click the small numbers 1 and 2, respectively.
41. To summarize, there is a 95% match for the human target sequence on exon 2 of the monkey TTR gene, and a 95% match for the monkey target sequence on exon 1 of the human TTR gene. The image below can be saved for your reference. Click on the image to enlarge it, then right-click on the enlarged image to save it.
The above diagram only shows one strand of DNA matched against a target sequence. The corresponding double-stranded DNA configurations are shown below, for exon 1 (top) and exon 2 (bottom). Note that the PAM can be located either on the template strand (exon 1) or the coding strand (exon 2). Regardless of which strand it is on, the PAM is always downstream (towards the 3′ end) of the target sequence on that strand*.
*Note: Comparison of the double stranded sequences above shows why the reverse complement of Gillmore’s monkey target sequence was used to search the human and monkey TTR gene sequences. The file DNA_TTR_gene_monkey.txt was the reverse complement of the original TTR gene sequence from Genbank, so it had to be searched using the reverse complement of Gillmore’s target site, shown below from Fig.3B of that publication. Doing this enabled the coding strands of both the monkey and human genes to be searched for mismatches, as shown above (letters highlighted in blue are on the coding strand). 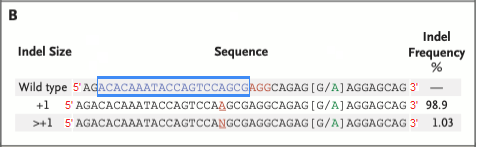
For their monkey trials, Gillmore’s team chose a guide RNA that required a PAM on the template strand, but for their human trials they chose a guide RNA that required a PAM on the coding strand. This choice of coding versus template strand does not affect Cas9 function, but the fact that they targeted different exons for monkeys and humans does raise some interesting questions (see below).
Questions
(1) For both exon2 and exon1, the mismatch is located relatively close to the PAM (4 bp away for exon 2, and right next to the PAM for exon 1). Is it possible for Cas9 to cut when mismatches are located this close to the PAM?
(2) How might Gillmore’s team have dealt with this, if they wanted to use the similar guide RNAs for both the human and primate trials?
(3) Which would give better results, using different guide RNAs that target different exons with a 100% match, or using 95% similar guide RNAs that target the same exon?
(4) Here is a link to statements from Gillmore’s protocol giving the reason for their decision to use separate guide RNAs for the human and monkey trials, and another link describing why crab-eating macaques were used as primate surrogates (‘NHP’ in the statements stands for ‘non-human primate’). Is there any conflict between these two statements? .
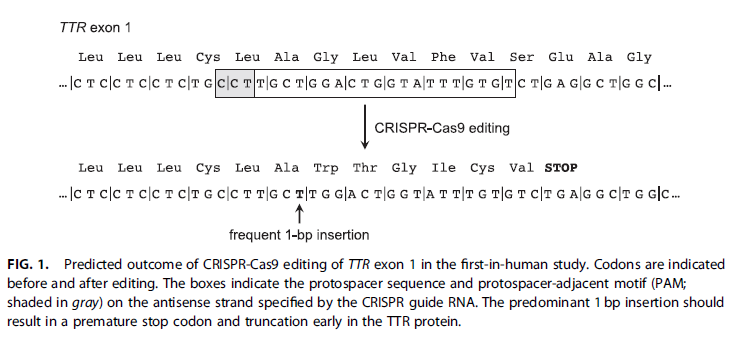
(5) The quote and figure below are from a news article in the CRISPR Journal about Gillmore’s study (“CRISPR Hits Home in a First-in-Human study”,Musunuru 2021). The author of this article had not read Gillmore et al. 2021 carefully, and made a major assumption that was in error. What was that assumption? Hint: Compare Fig 1. below with the data in Step 41 above, and also with the diagram in Step 23.
“These promising results paved the way for the monkey studies and human patient studies reported in the recent New England Journal of Medicine report.3 The investigators chose a guide RNA that targets a sequence in TTR exon 1 and demonstrated that it achieved highly efficient editing in hepatocytes. With the goal being to introduce disruptive insertion or deletion (indel) mutations via non-homologous end joining, which typically yields a broad variety of semi-random indels, the tested TTR guide RNA yielded the same 1 bp insertion ∼99% of the time—an unusually high consistency more typical of base editing. This insertion is predicted to result in a frameshift mutation with a premature stop codon several positions downstream of the edited site, a reasonably clean truncation of the TTR protein (Fig. 1).”
Top of this page
Go to Part 1: Overview of ATTR, CRISPR, and Gillmore et al. 2021
Go to Part 2: Cut the TTR gene with Cas9 under the direction of guide RNA
Go to Part 3: Look for potential off-target effects of Cas9
Go to Part 4: Examine mutations of patients associated with the study