Goal of this exercise: Analyze the first study that used the CRISPR Cas9 system to successfully knock out an abnormal gene in human patients (Gillmore et al. 2021). Part 4 will examine mutations in the patients in this study, using databases and viewers at the NCBI site.
Part 1: Overview of ATTR, CRISPR, and Gillmore et al. 2021
Part 2: Cut the TTR gene with Cas9 under the direction of guide RNA
Part 3: Look for potential off-target effects of Cas9
Part 4: Examine mutations of patients associated with the study
Part 5: Analyze frameshift mutations in monkeys and humans
Overview of Part 4
In Part 4 the TTR mutations from six patients will be examined using Case It v7.0.4 and NCBI tools including the SNP and ClinVar databases and Variation Viewer. The following quote is from Gillmore et al. 2021:
“The patients were 46 to 64 years of age, and four of the six patients were men; the body weight ranged from 70 to 90 kg. Three patients had a p.T80A mutation, two a p.S97Y mutation, and one a p.H110D mutation.”
Organization of Part 4
Steps 2-11: Use Case It v7.0.4 to identify mutations on TTR gene sequence.
Steps 12-19 : Use NCBI SNP and ClinVar databases to find information about the mutations.
Steps 20-26: Use NCBI Variation Viewer to visualize mutations on chromosome 18.
1. Open or activate Case It v7.0.4. If you do not already have Case It v7.0.4, go to Step 1 of Part 2 for instructions on acquiring the software. If you are using Case It on a Mac, read these instructions for accessing files.
IDENTIFY MUTATIONS ON THE TTR GENE SEQUENCE
Goal of Steps 2-11: In the following steps we will use the sequences of three known mutant sequences, determined from sequencing the TTR gene of several patients, to search the genome for the position in the genome of these mutations. Each mutant will be used as a ‘probe’ to scan the genome for matching sequences. Because the mutant sequences will not match the wild-type sequences exactly (because they are mutant), we first need to change the requirements of the search in order to find sequences that match at a level of 95%.
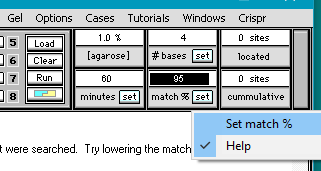
2. Double-click in the match % field and type in the number 95.
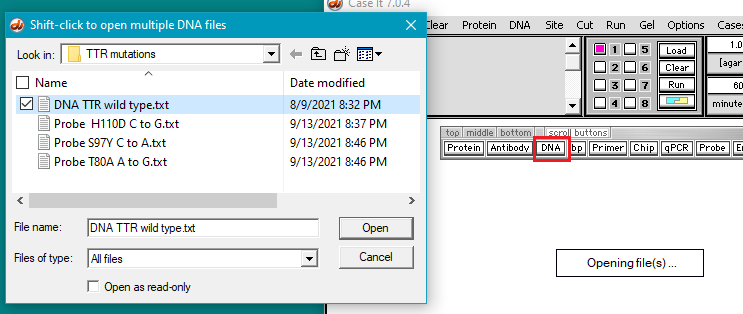
3. The DNA sequence and probes used for this part of the experiment can be found in the TTR mutations folder in the Case It v704 PC > TTR > TTR mutations folder. Click the DNA button and open DNA TTR wild type.txt. This file contains the sequence of the wild-type TTR gene. In this case we are only using the TTR gene itself rather than a larger portion of chromosome 18 as in previous experiments. We will search this sequence to find the position of the three mutations mentioned above.
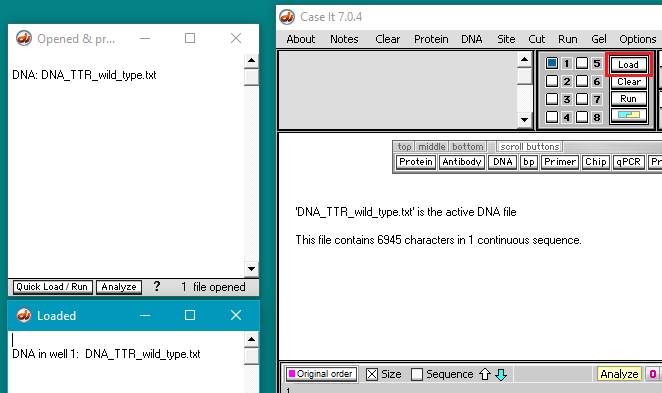
4. Click the Load button to load the DNA sequence into well 1. The well must be loaded for the DNA sequence to be searched in the following steps. This is just the way the program works, as we will not be running a gel.
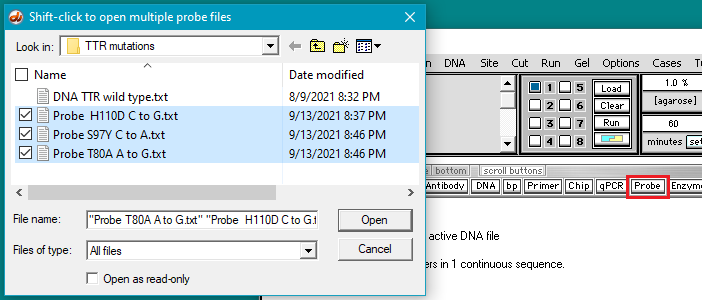
5. Click the Probe button and shift-click to open three files containing the mutation sequences.
Note: Many molecular biology techniques involve detecting specific DNA sequences using probes. A probe is a short DNA sequence which shares sequence similarity, or homology, with the target sequence. In lab based experiments, probes are labeled (e.g. with a radioactive isotope) and the hybridized to target DNA and visualized based on the presence of the label. Computer based experiments work in a similar way. A short DNA sequence is used as a ‘probe’ and used to search much larger DNA sequence for regions that match the probe sequence.
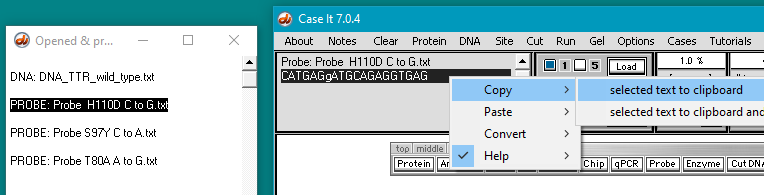
6. The three probes appear in the Opened & processed (O&P) window on the left. Follow this three-step procedure:
(1) Click on the first probe listed in the O&P window. The probe sequence will appear in the gray field on the main screen, as shown below.
(2) Double-click on the probe sequence in the gray field to highlight it.
(3) Right-click in the gray field and select Copy > selected text to clipboard.
Note: In this probe’s name, H110D refers to the mutation at the amino acid level, in which a histidine (H) is changed to an aspartic acid (D). C to G refers to the change at the DNA level.
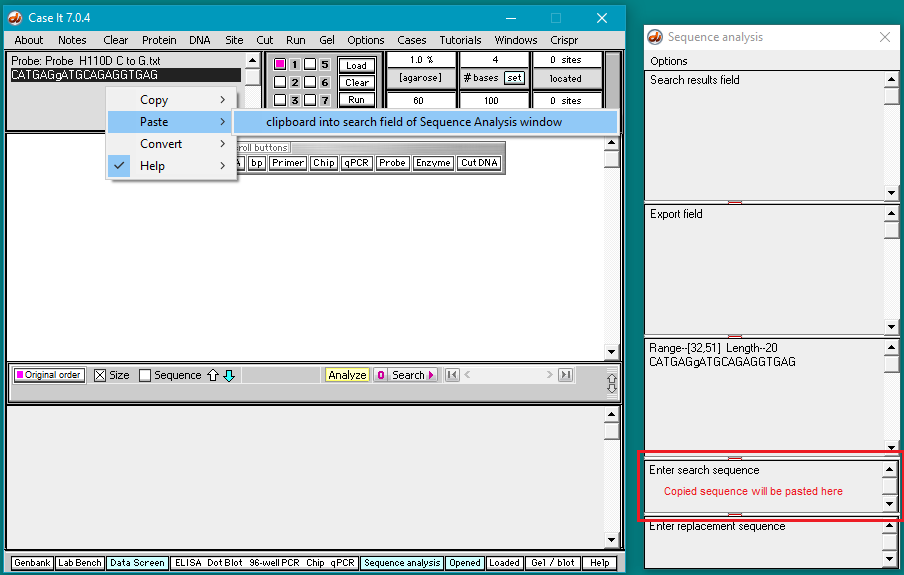
7. Right-click again and select Paste > clipboard into search field of Sequence Analysis window. The copied sequence will be automatically pasted into the field at lower right as shown below.
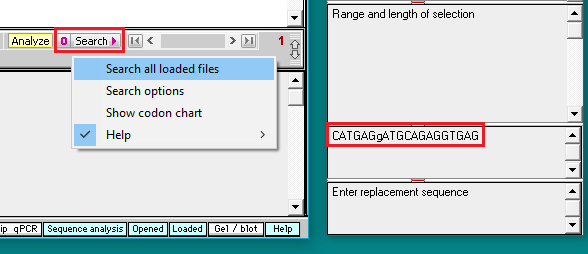
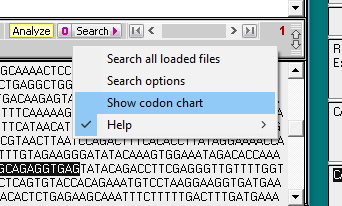
8. Click on the Search button and select Search all loaded files.
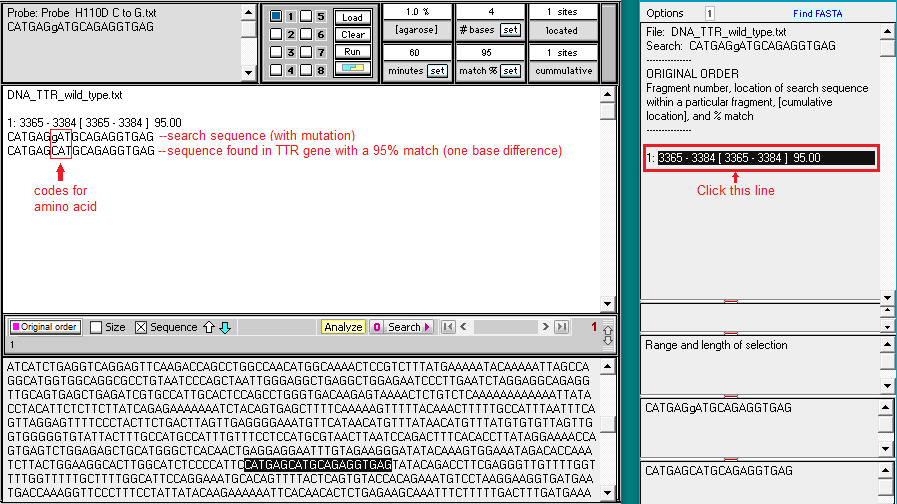
9. Search results appear in the top field of the Sequence Analysis window, and also in the top field of the main window. Click on the line in the top field of the Sequence Analysis window and the location of the search sequence in the TTR gene is highlighted in the lower field of the main window. Note the difference between the search sequence and the sequence located on the TTR gene sequence – one of the 20 bases is different, since the search sequence had the H110D mutation. This is the reason why the match % had to be set at 95. If we had left it at the default value of 100%, the location would not have been found.
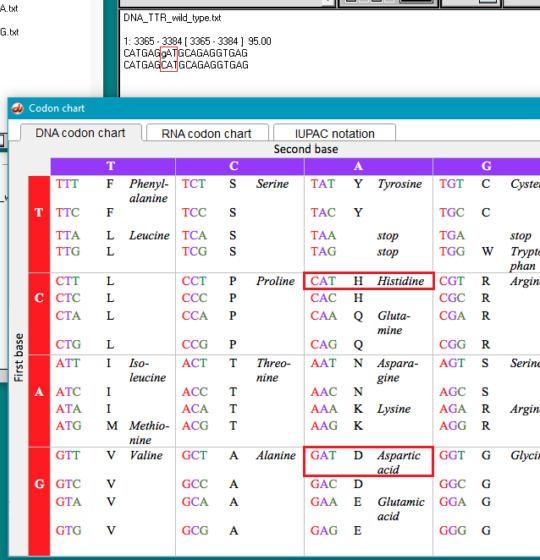
10. Click on the Search button again, but this time select Show codon chart.
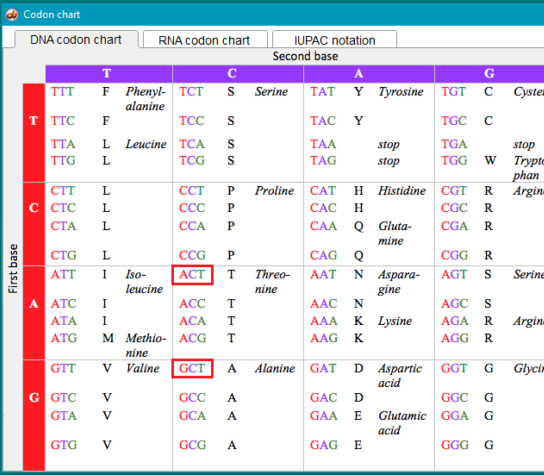
11. For the wild type TTR sequence, CAT codes for the amino acid histidine (symbol H), and GAT codes for aspartic acid (symbol D). For the normal protein coded by the TTR gene, histidine is found at position 110, whereas for the mutated protein, aspartic acid would replace histidine at this position. This is the reason why the mutation is called p.H110D, with p standing for ‘protein’.
Exercise: Use this same procedure to identify the p.S97Y and T80A mutations.
FIND INFORMATION ABOUT THE MUTATIONS
The goal of the next series of steps (12-19) is to use the NCBI databases to find more information about the mutations.
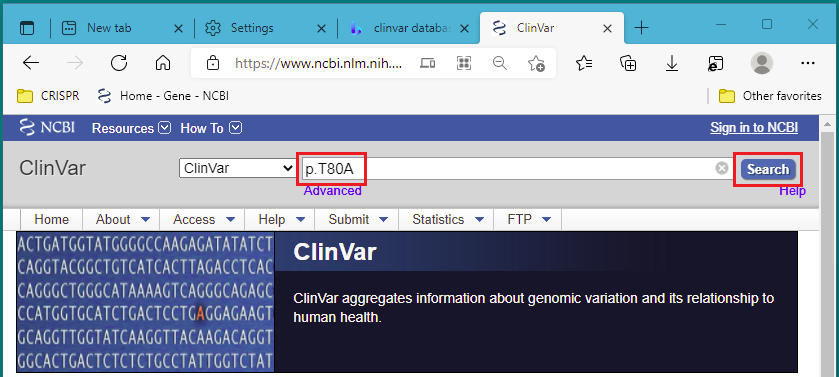
12. Right-click on this link to open the NCBI ClinVar database in a new tab or a new window. Start with the first mutation type, p.T80A. Paste or type p.T80A into the search field, and click Search.
Note: From the ‘about’ link of the ClinVar: “ClinVar is a freely accessible, public archive of reports of the relationships among human variations and phenotypes”. This means that ClinVar is a collection of clinically relevant genetic variations.
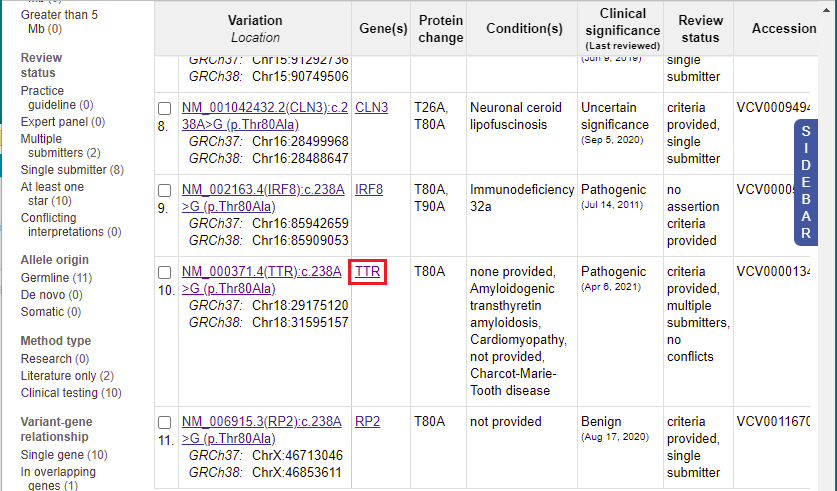
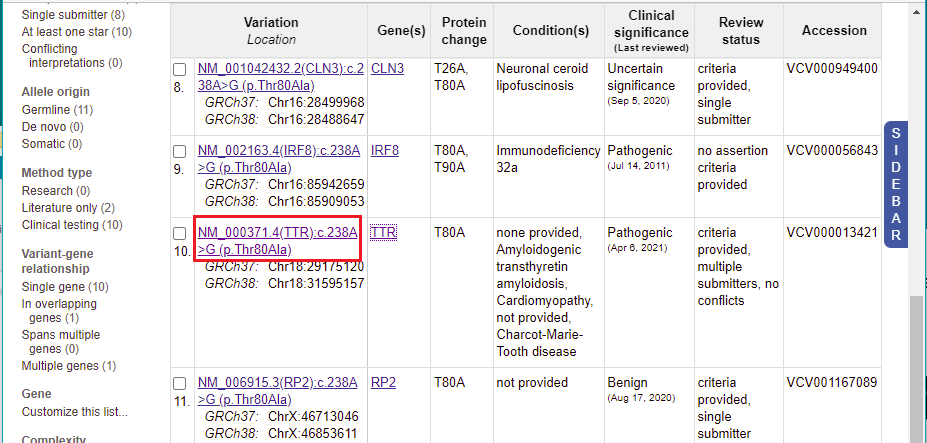
13. A list of 11 entries appears for this mutation. Scroll to the 10th entry, then click on ‘TTR’ in the Gene(s) column.
14. You are taken to the Gene database, which provides extensive information on the the TTR gene, accessed via links to other databases with information on this gene. Scroll down this page to show how much information is given about the gene. You can explore this information at a later time, but for now return to Clinvar by clicking its tab or window.
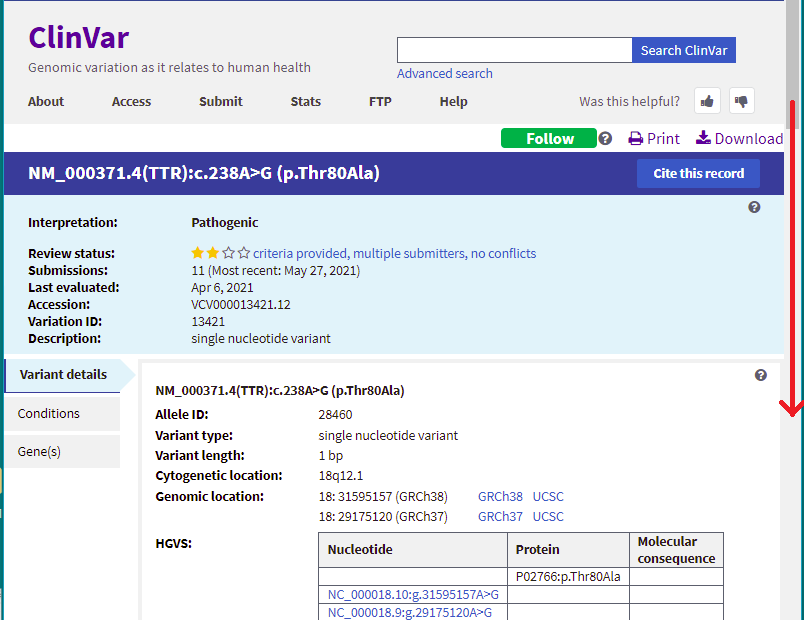
15. You are back on the ClinVar page. Click the link shown in the Variation column for the 10th entry.
16. Scroll down on this page to see a list ‘Submitted interpretations and evidence’ for this gene, along with a list of ‘Citations for this variant’ with links to the PubMed database. This is a very valuable source of information on research into conditions caused by mutations in the TTR gene, that you can explore at a later time.
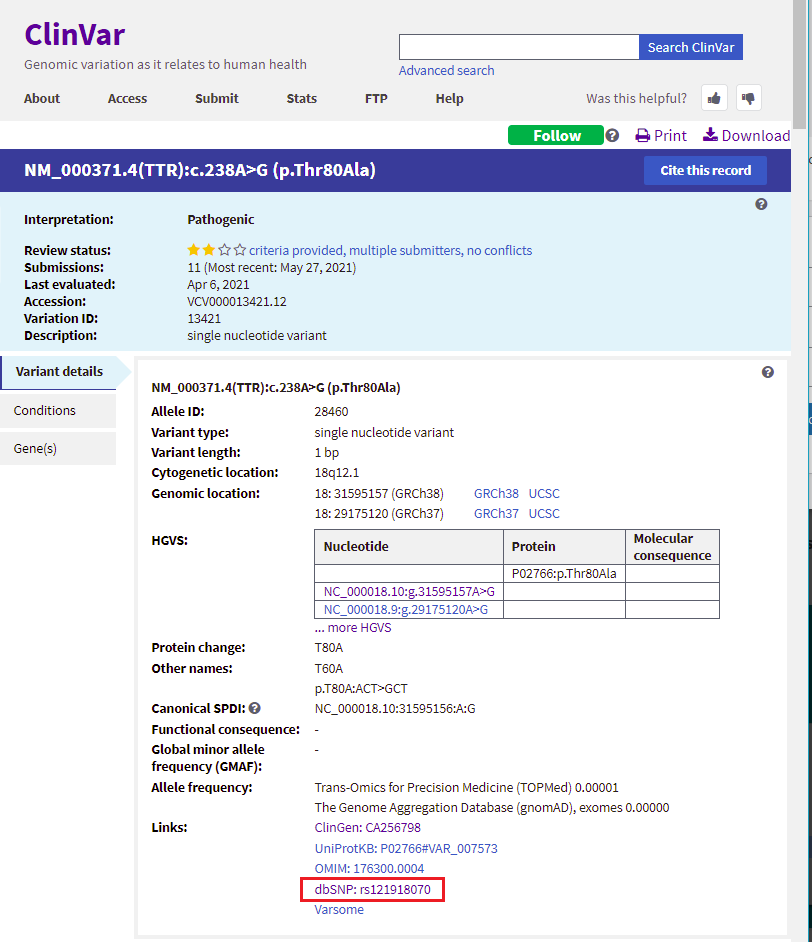
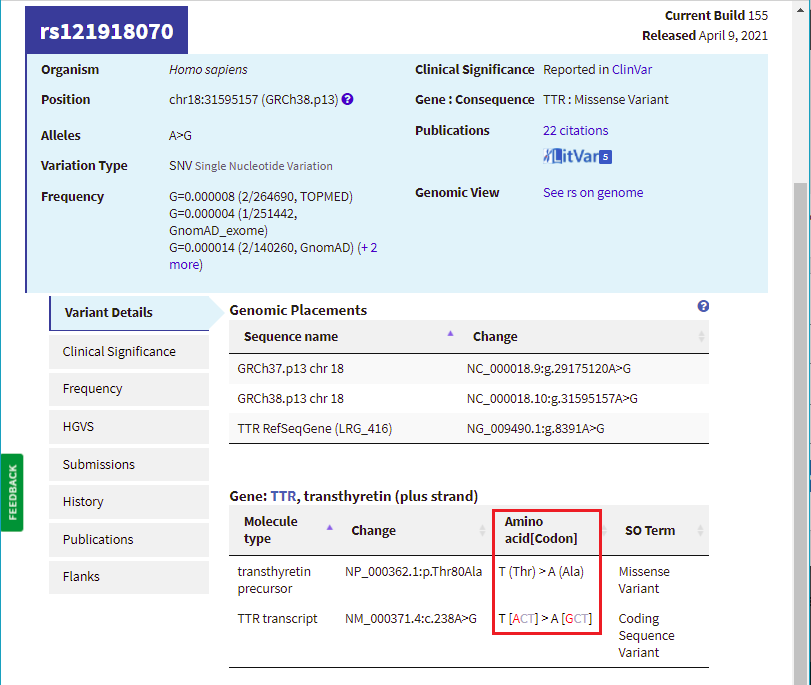
17. Scroll back up on the page and click the dbSNP: re121918070 link (red box below) to go to this entry in the SNP database.
18. You are now in the NCBI SNP database. Information on this page shows the nature of the mutation (the amino acid threonine changed to alanine, because of a change in the DNA sequence from ACT to GCT. Leave your web browser open to this page, as you will be referencing it again in Step 20.
Note: NCBI is the National Center for Biotechnology Information. This center houses many useful databases and bioinformatics tools for genome analysis. The SNP database is a collection of all known single nucleotide polymorphisms (which means single nucleotide differences) known in the human genome. Searching this database using our mutations of interest allows us to find previous information known about these specific changes.
19. Go back to Case It v7.0.4. Click the Search button, and select Show codon chart.
The mutation change from threonine to alanine is shown in the red boxes, along with the other codon combinations associated with these amino acids.
VISUALIZE MUTATIONS USING VARIATION VIEWER
The goal of the next series of steps (20-25) is to use the NCBI Variation Viewer to visualize DNA sequences associated with the mutations and determine exon locations for these mutations.
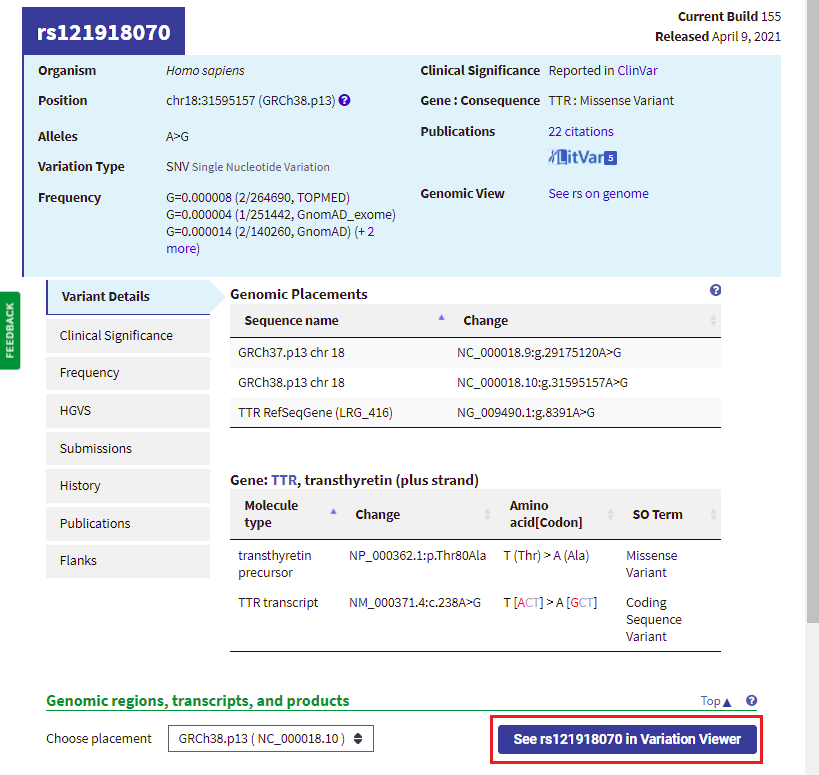
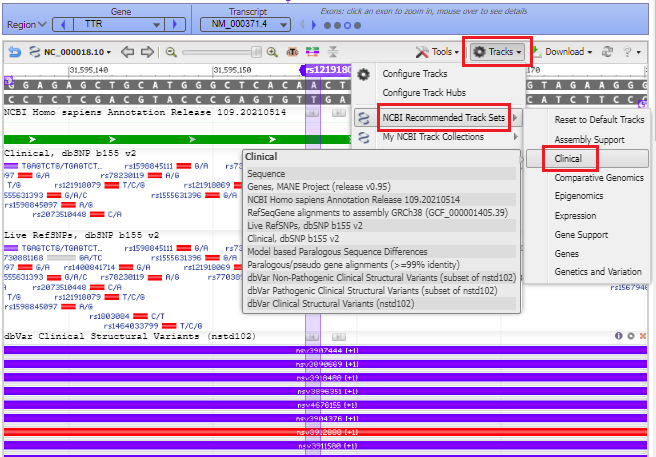
20. Your web browser should still be on the same page that it was on in Step 18. Click the button labeled ‘See rs121918070 in Variation Viewer‘ (shown in red box below).
21. A series of purple and red lines appear, indicating structural variants. Click on the Tracks button and select NCBI Recommended Track Sets > Clinical from the drop-down menu.
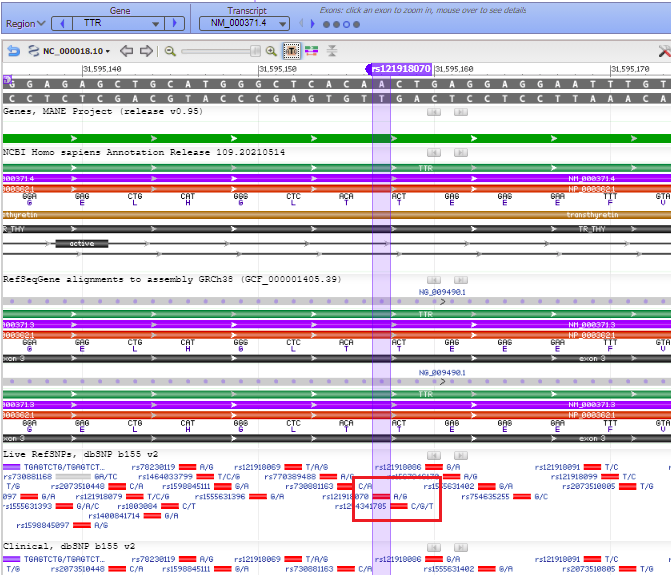
22. The purple marker lines up with a small solid red box with A/G next to it, indicating the change from A to G if the mutation is present (see Step 18 above).
23. Hover your mouse over the colored bars to see pop-up descriptive windows. For example, hovering over the solid red A/G box gives links to databases. The red bar represents the precursor to the TTR protein, showing DNA base triplets associated with each amino acid.
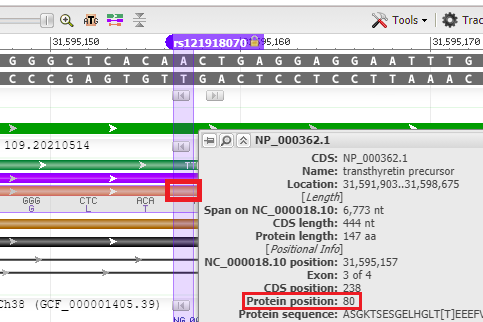
24. Hover your mouse over the red bar for the protein, and the position of the amino acid is shown (80th amino acid from the beginning of the protein). Recall that the name of this SNP is ‘T80A’, meaning threonine (T) changed to alanine (A) at the 80th amino acid position. The SNP is located on exon 3 of the TTR gene.
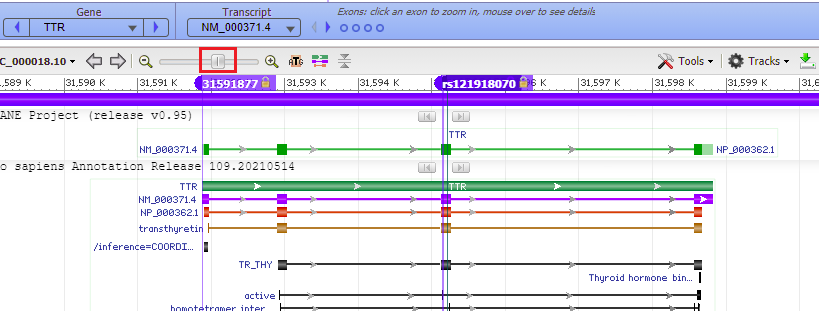
25. Move the slider to the left to zoom out to see the entire TTR gene, to verify that the mutation is located on exon 3 (slider is positioned at 60%, if you hover over the slider knob). Recall that the guide RNA used by Gillmore’s team caused Cas9 to cut the gene at exon 2 (Step 41 in Part 2). Click on the link to go to Step 41 of Part 2, then use the back arrow of your browser to return here.
Question: The guide RNA/Cas9 complex that Gillmore’s team used cut the gene in exon 2. Would a mutation that is located on exon 3 be disabled by a cut in exon 2?
26. Repeat the above procedure for the p.S97Y mutation, starting with the SNP database. The p.H110D mutation was apparently not entered into the SNP or Clinvar databases, but is cited in PubMed.
Top of this page
Go to Part 1: Overview of ATTR, CRISPR, and Gillmore et al. 2021
Go to Part 2: Cut the TTR gene with Cas9 under the direction of guide RNA
Go to Part 3: Look for potential off-target effects of Cas9
Go to Part 5: Analyze frameshift mutations in monkeys and humans