Background: Cases A, B and C deal with PKU, a genetic metabolic disease caused by a mutation in the phenylalanine hydroxylase enzyme. In the most common form of PKU, a C to T point mutation (SNP) causes an arginine to be replaced by tryptophan at amino acid position 408, resulting in an inactive enzyme and incomplete metabolism of phenylalanine-containing compounds such as proteins. The NCBI designator for this SNP is rs5030859 (used in Step 1 below). There are a variety of ways to obtain information about this SNP using NCBI tools, three of which will be demonstrated below (Genome Data Viewer, dbSNP, and Structure).
Goal of Steps 1-7: Use the Genome Data Viewer and SNP database to learn more about SNPs associated with PKU.
Goal of Steps 8-15: Use the NCBI Structure tool to see the replacement of arginine by tryptophan in 3 dimensions.
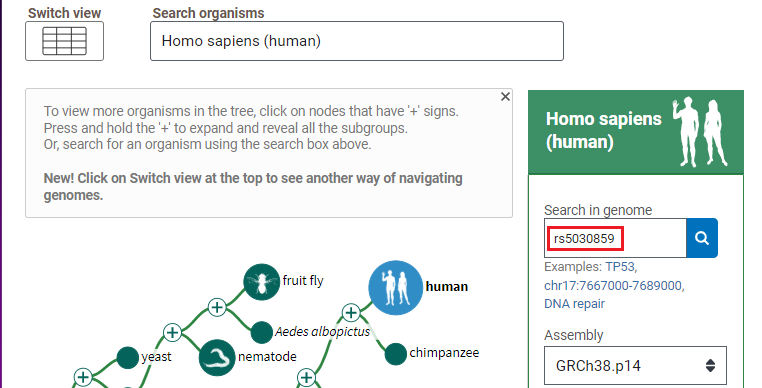
1. Open the Genome Data Viewer and enter rs5030859 into the search box, as shown below, and click the magnifying glass icon so search for this SNP. If you copy and paste, use control-V or command -V to paste.
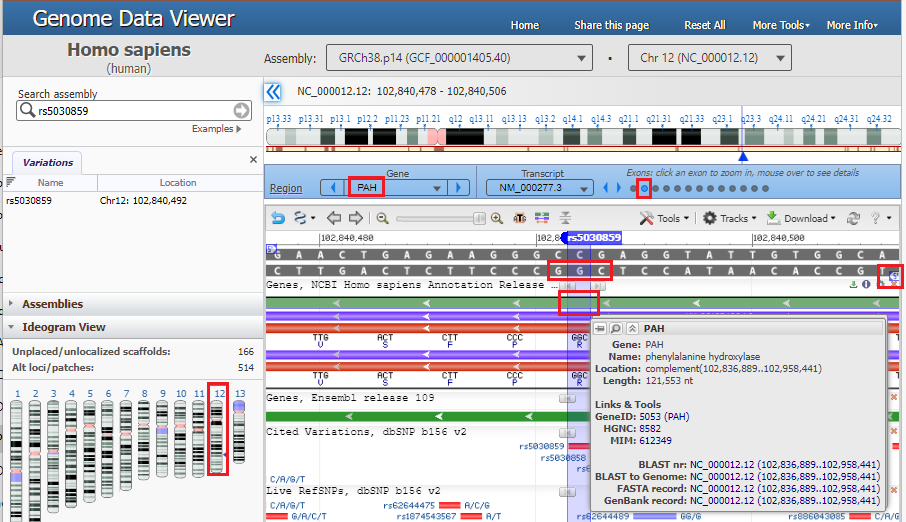
2. The location of the SNP is shown on the gray double-stranded DNA sequence, which is also represented by the top green line. Click on the top green line and the amino acid sequences of the protein will appear, represented by the red line. The direction of arrows point from right to left, indicating the 5′ to 3′ direction of the DNA molecule, so the nucleotides in the red box should be read from right to left as “CGG”, a codon for arginine (R). Hover your mouse over the top green line, in the location of the red box on that line, and a pop-up box will appear giving the name of the gene (PAH) and its associated protein (phenylalanine hydroxylase). There are 13 small circles in the blue bar, representing the 13 exons of the PAH gene. The 12th circle from the left is open, indicating that this DNA sequence is part of exon 12.
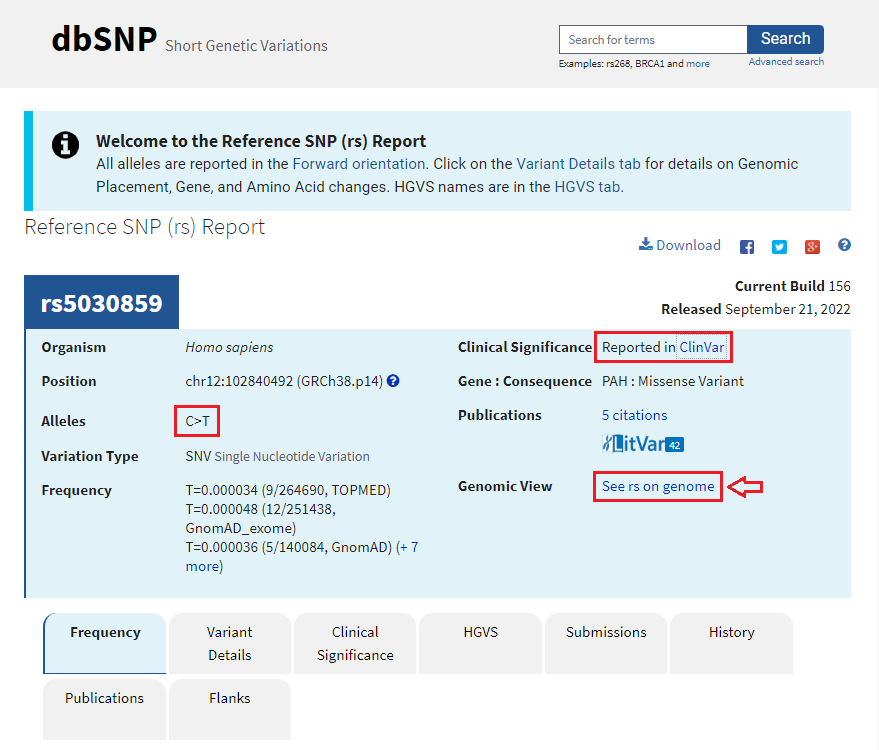
3. Move your mouse cursor down and hover over the red box labelled rs5030859, as shown below, and click on the rs5030859 link next to SNP summary in the pop-up box.
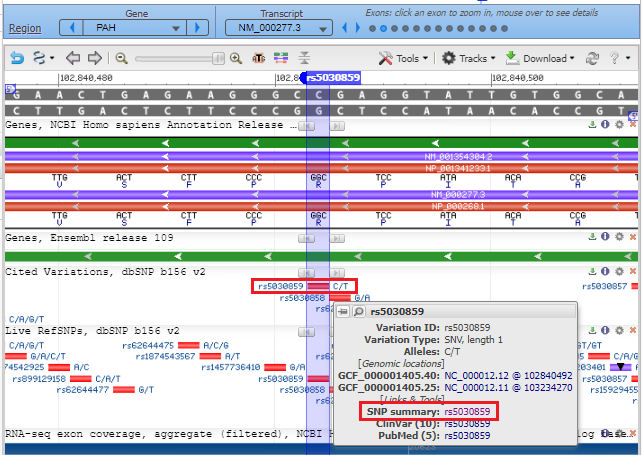
4. A new screen will appear in the box, with a scrollbar, with another link SNP summary rs5030859. Click on that link.
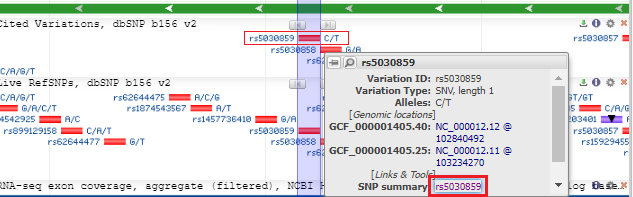
5. The SNP database entry will appear, indicating that the change is from C to T (CGG to TGG). Much information concerning this SNP can be found by clicking the various tabs and links on this page. Click See rs on genome to jump down to the bottom of the page.
Note: The information below can also be obtained by searching for rs5030859 in the dbSNP database.
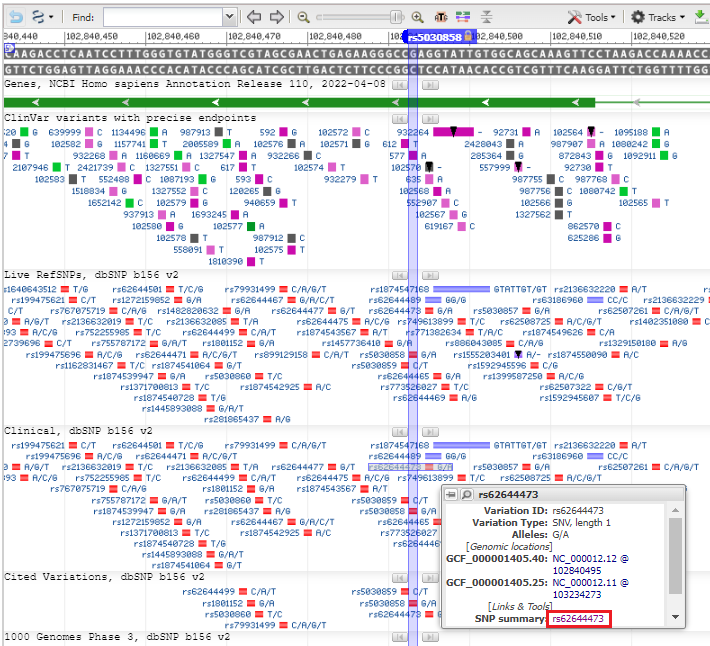
6. A view appears similar to that of the Genome Data Viewer, showing the same SNP location found earlier, along with all of the SNPs associated with this exon. Experiment by hovering over different SNPs close to rs5030858, and then hover over SNP rs62644473 and click on the SNP summary link in the pop-up box.
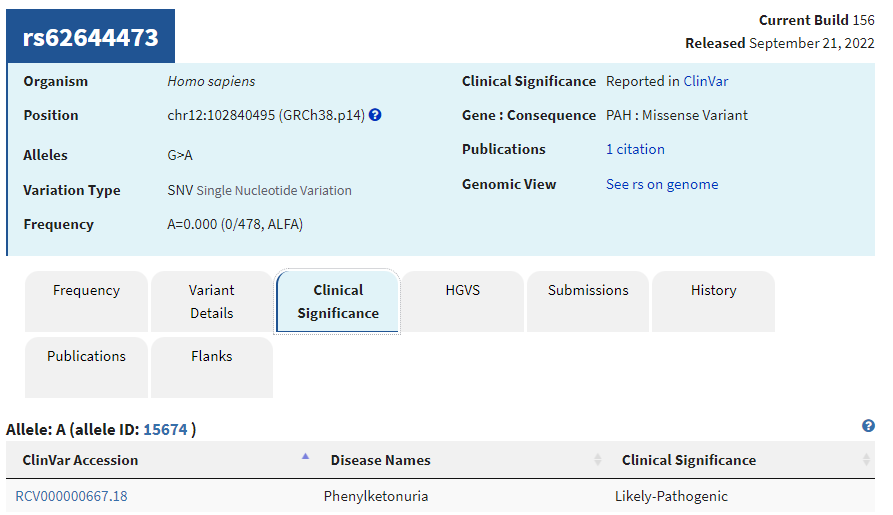
7. Click on the Clinical Significance tab and note that this SNP is also associated with phenylkentonuria (PKU).
Question: How many other SNPs can you find associated with PKU or other clinical conditions?
Goal of Steps 8-15: Use the NCBI Structure tool to see the replacement of arginine by tryptophan in 3 dimensions.
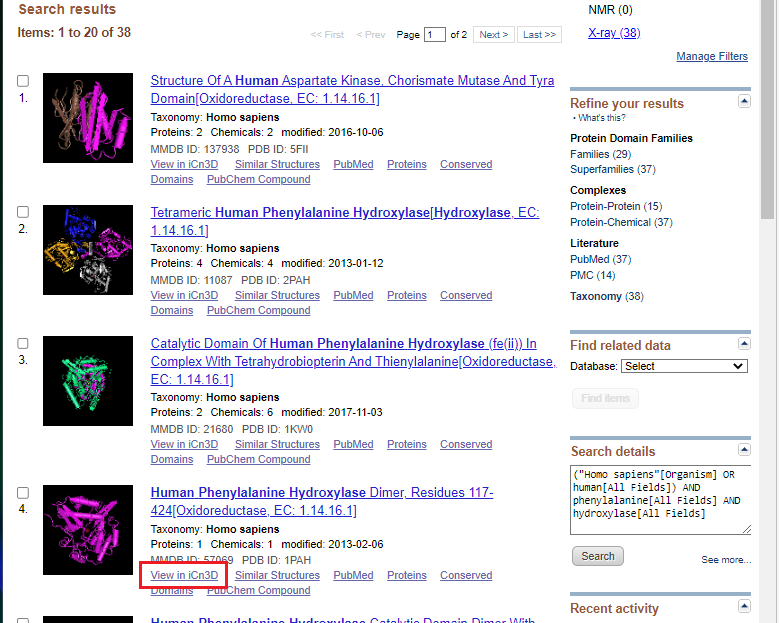
8. Open NCBI Structure, enter human phenylalanine hydroxylase into the search box, and click Search. If you copy and paste, use control-V or command -V to paste.
9. Click the view in iCn3D link as shown in the red box below.
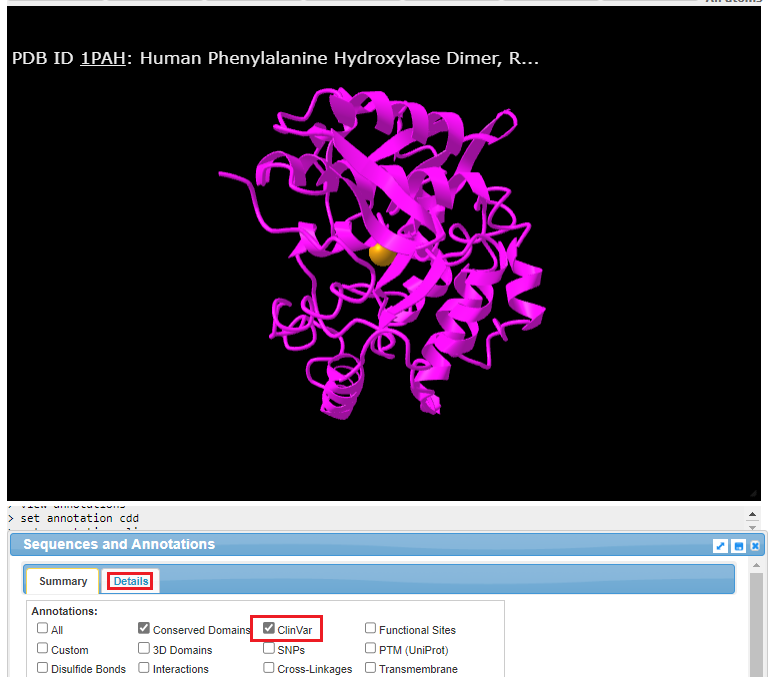
10. The structure can be rotated in three dimensions using your mouse cursor and wheel, and the entire structure can be moved using by right-clicking and dragging. Click the ClinVar checkbox, then click the Details tab.
Note: On a tablet or phone, use one finger to rotate the structure, and two fingers to move the entire structure. If the structure disappears when you attempt to manipulate it, hit the back arrow of your browser to go back to Step 9, and click on View in iCn3D again. You may have to experiment with finger movements, especially on a phone.
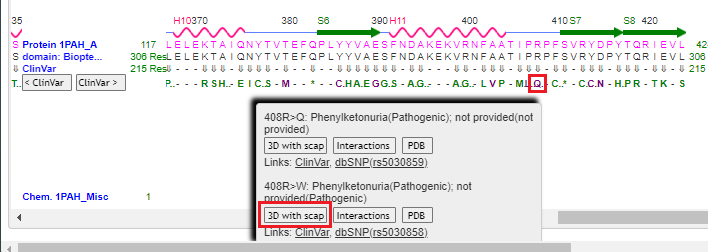
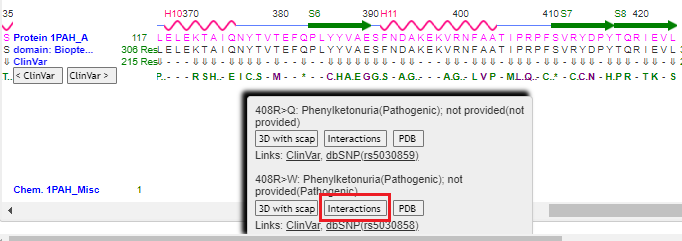
11. Drag the scrollbar to the right to see position 408 on the protein, and hover over the Q using your mouse cursor. Click on the 3D with scap button. Note: On a phone or tablet, carefully drag the protein sequence with your figure to see position 408, on the far-right end of the protein. Delicately touch the Q with your finger until the pop-up box appears.
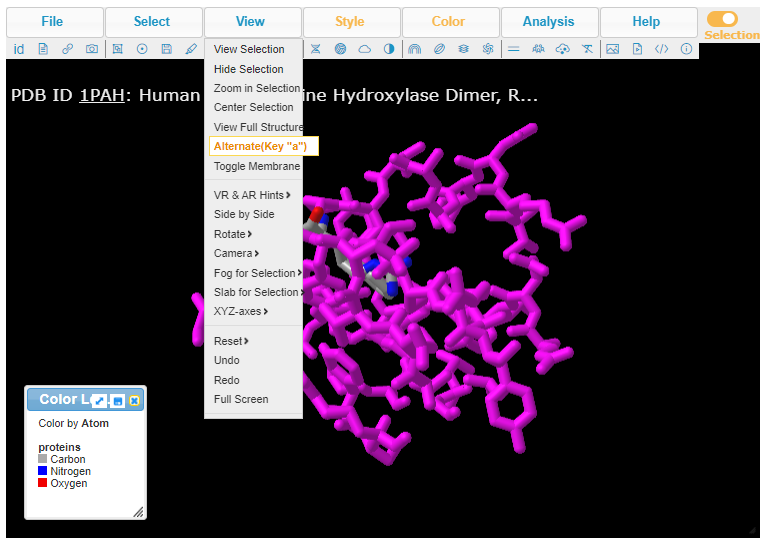
12. Press the A key on your keyboard (if using a computer) or use the View menu (if using a tablet or phone). Either method will allow you to alternate between seeing arginine or tryptophan at position 408, in three dimensions. Note: On a phone, the View menu can be accessed via a menu button with three lines at upper left, not shown here.
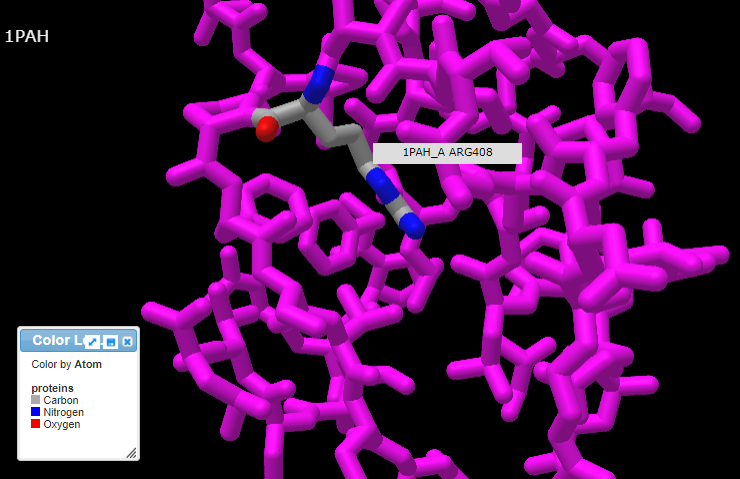
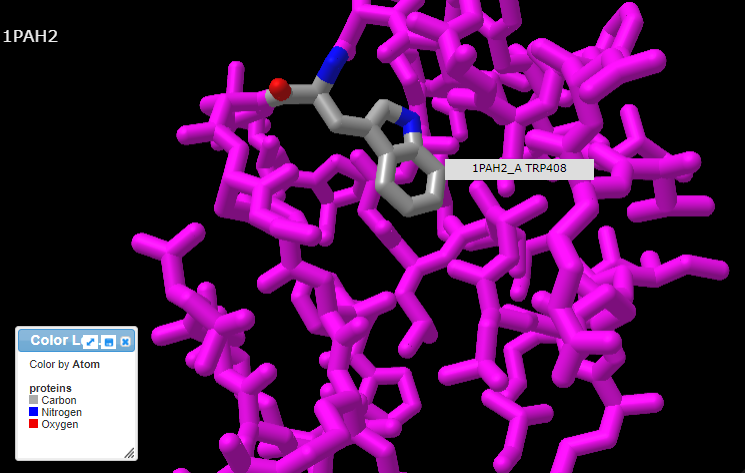
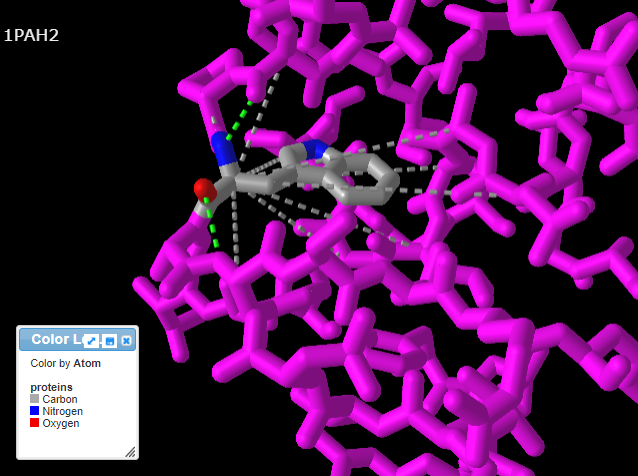
13. Pressing the A key or using the View menu will allow you to toggle between arginine (shown directly below) and tryptophan (second image below). Rotate the structure by dragging with your mouse, and enlarge using your mouse wheel (or the pinch method on a phone or tablet). The label designations shown below refer to arginine or tryptophan at position 408 of the protein. Click here to see a video showing how the structure can be rotated to identify the adjacent amino acids (proline in both cases).
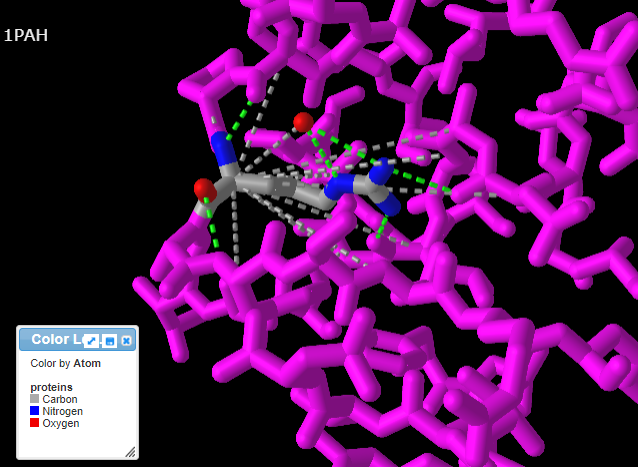
14. Repeat steps 9-11 above, but this time click the Interactions button in the pop-up box. Note: If you do not repeat steps 9-11, you will get an error message.
15. Use your mouse and mouse wheel (or the pinching method) to rotate and enlarge the protein to see the interactions between the amino acids and neighboring amino acids. Press the A key or using the View menu will allow you to toggle between arginine (shown directly below) and tryptophan (second image below). Note: The extra ‘floating’ oxygen is an anomaly in the model.
Questions:
1. How might changes in these interactions interfere with protein (enzyme) function?
2. What is the function of phenylalanine hydroxylase, and why is it so important that the body be able to metabolize phenylalanine-containing compounds?