Background: Genetic mutations such as those in Cases A and B are the cause of early onset (familial) Alzheimer’s disease, comprising only a small percentage of cases worldwide. The vast majority of Alzheimer’s cases are caused by a combination of factors occurring most often in people over 65, a condition called late-onset Alzheimer’s disease (LOAD). Extensive (and some suspect) research on these factors has yet to identify the precise cause of LOAD, although lifestyle, environmental, and genetic risk factors may all play a part. This section will examine one genetic factor that may be correlated with an increased risk of Alzheimer’s disease, involving an allele of the gene for apolipoprotein E. The gene that codes for this protein is called APOE.
Goal of Steps 1 – 14: Examine two SNPs associated with allele variation of the APOE gene.
Goal of Steps 15 – 24: Examine wild-type and mutant variations on the three-dimensional structure of the APOE protein.
Goal of Step 25: Examine a pioneering research article that used base editing to correct a SNP mutation in the APOE gene.
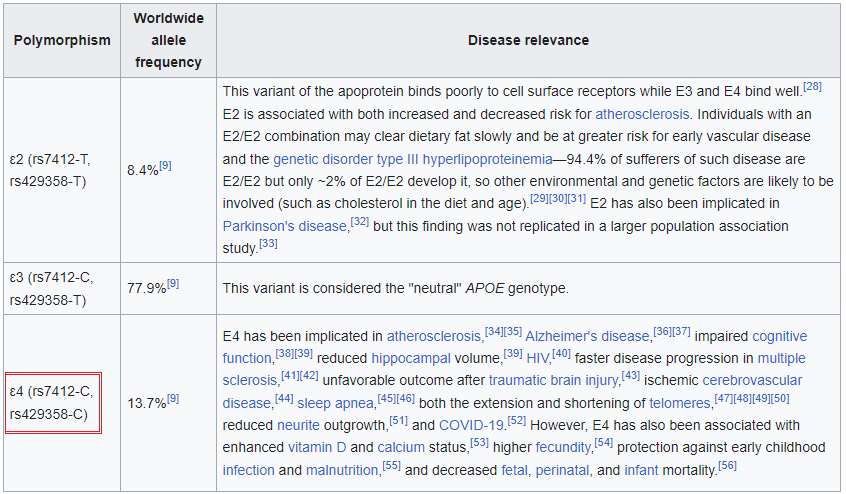
1. Click on this link to open the Wikipedia page to the entry for apolipoprotein E and read about the function of this protein. Scroll down on that page to a table that describes the three alleles for the APOE gene that codes for this protein (reproduced below). Note that the gene exhibits ‘polymorphism’, meaning that three different alleles are present in the human population: APOE2, APOE3, and APOE4, designated as e2, e3, and e4 in the table below. These alleles differ in the nucleotides present at two SNP locations: APOE2 has thymine (T) present at both locations, APOE3 has a cytosine (C) present at one location and a T present at the other, and APOE4 has C present at both locations. These SNP locations are designated as rs7412 and rs429358 in the NCBI database.
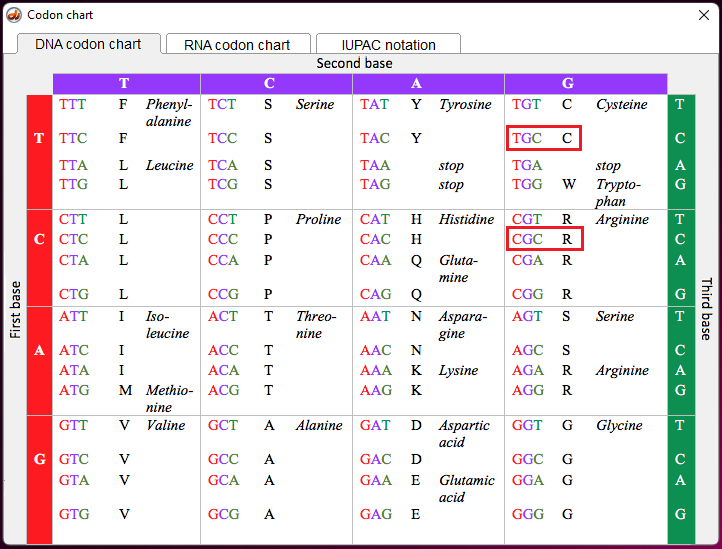
Note: The letters T and C in the above table (left column) refer to variations in nucleotides that are part of codons in the DNA sequence for the APOE gene. The codons associated with rs7412 and rs429358 are either TGC or CGC. For the purpose of this exercise, remember that TGC is a codon for the amino acid cysteine (also confusingly abbreviated C), and CGC is a codon for the amino acid arginine (R), as shown below (codon chart from Case It v705).
2. Click on this link to open your browser to the Gene web page of NCBI (National Center for Biotechnology Information), type APOE into the field as shown below, and select Homo sapiens APOE from the drop-down menu.
3. Click on the Genome Data Viewer button.
4. The APOE gene location appears on the chromosome 19 nucleotide sequence. Note that four exons are present (green boxes) separated by three introns (green lines). Purple boxes represents mRNA, and red boxes represents protein sequences.
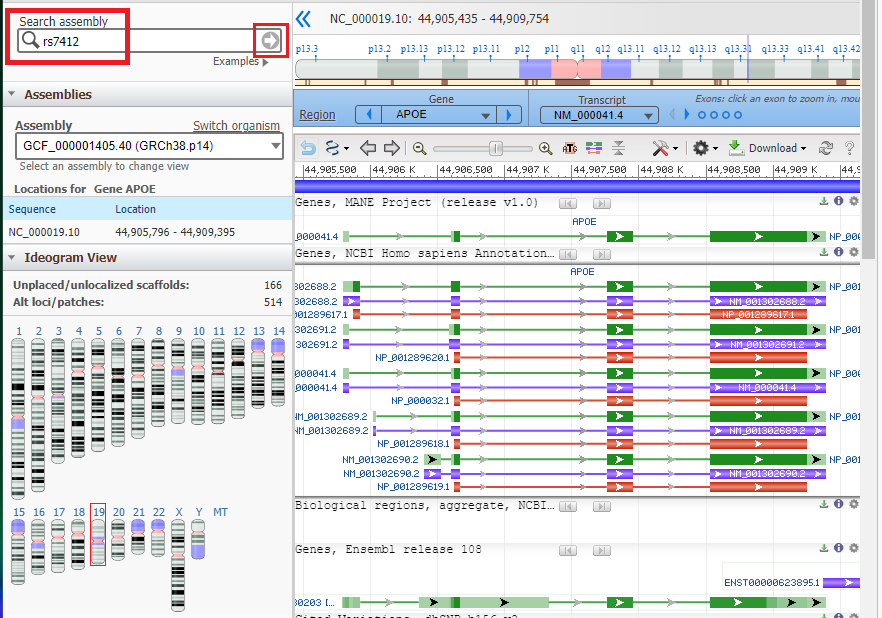
Type or (copy and paste) rs7412 into the Search Assembly box, then click the arrow in that box to search for that SNP. Note that this designation is case sensitive, so the ‘rs‘ should be lowercase.
5. The rs7412 SNP has been located on the APOE gene, as shown below. Note that cytosine (C) is present at this location on the top strand.
Question: Since C is present at the SNP location, this particular DNA sequence could be part of either the APOE3 or the APOE4 allele (see table in Step 1 above). How would you determine which of the two alleles this DNA sequence is from?
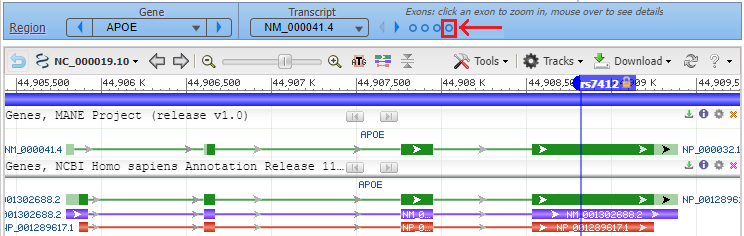
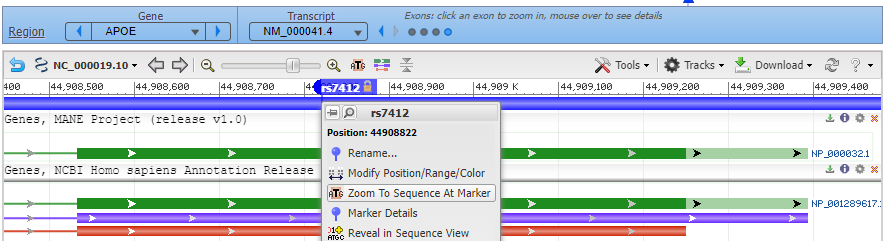
6. Click the back button on your browser to see all four exons, with the marker for rs7412 in the fourth exon. Hover your cursor over the fourth circle without clicking (red box), and the fourth exon will be highlighted. Then click on the fourth circle to show the entire sequence of the fourth exon (second image below).
7. Hover over the marker label and a box will appear as shown below. Select Zoom To Sequence At Marker to show the magnified view of the sequence at the marker location (as shown in Step 8).
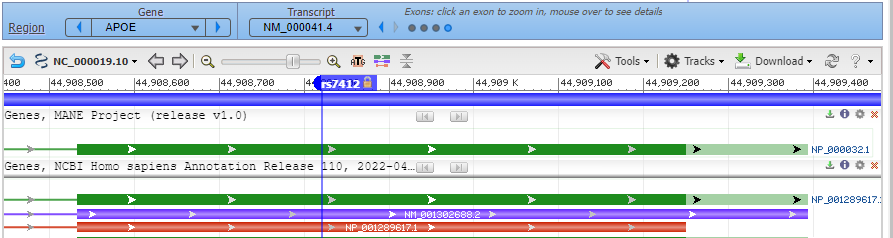
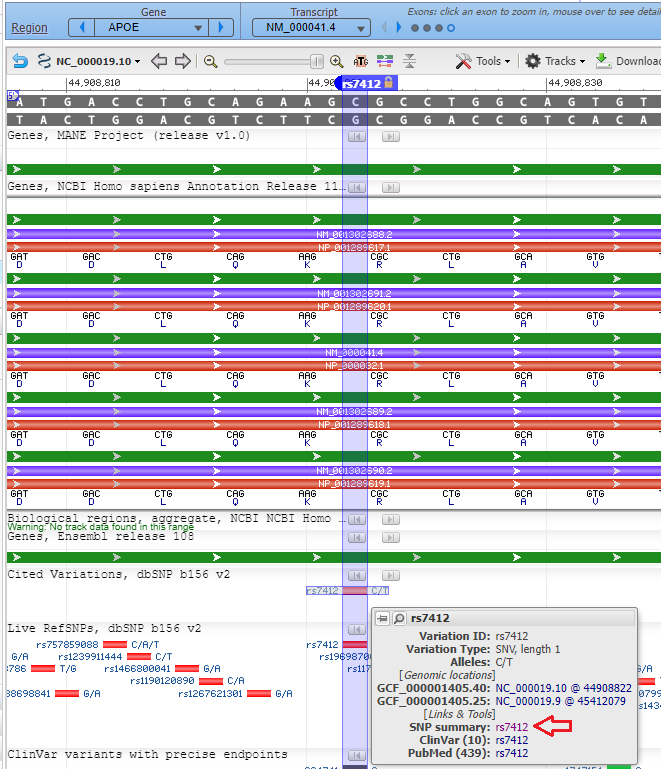
8. Click on the small red box labelled ‘rs7412’ and a box will appear as shown at the bottom of the graphic below. Then click ‘rs7412’ next to ‘SNP summary’, as shown at the red arrow.
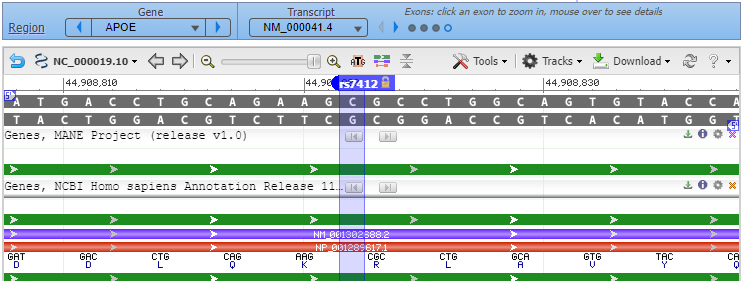
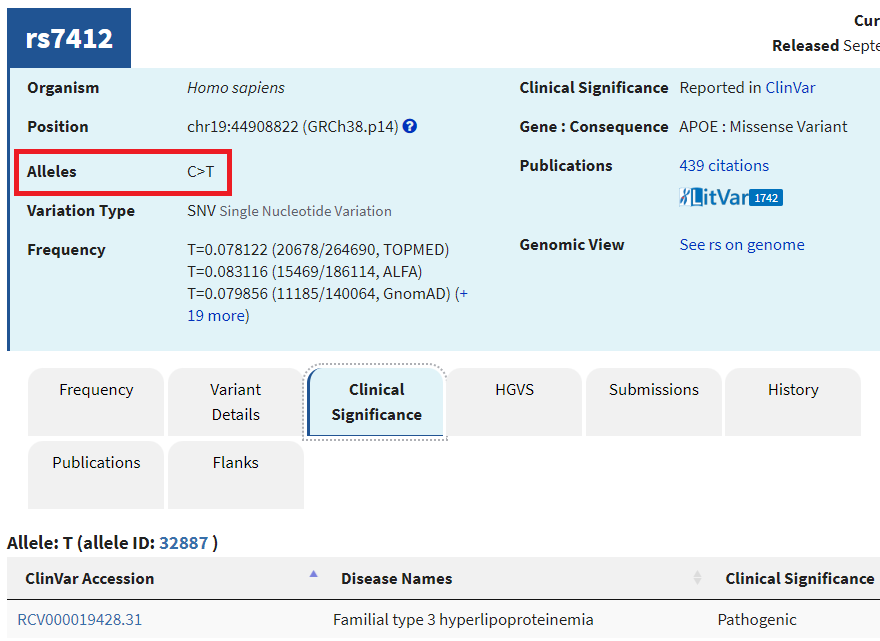
9. A new tab will appear on your browser showing this entry in the SNP database. C>T means that C can be changed to T at this location. Click on the Clinical Significance tab and note that the C > T mutation is associated with hyperlipoproteinemia.
Questions:
—Look again at the table in Step 1. Which allele of the APOE gene has T at this position, and which alleles have C at the position?
–Learn about hyperlipoproteinemia by clicking the link. Can you think of any possible relationships between this disease and Alzheimer’s disease?
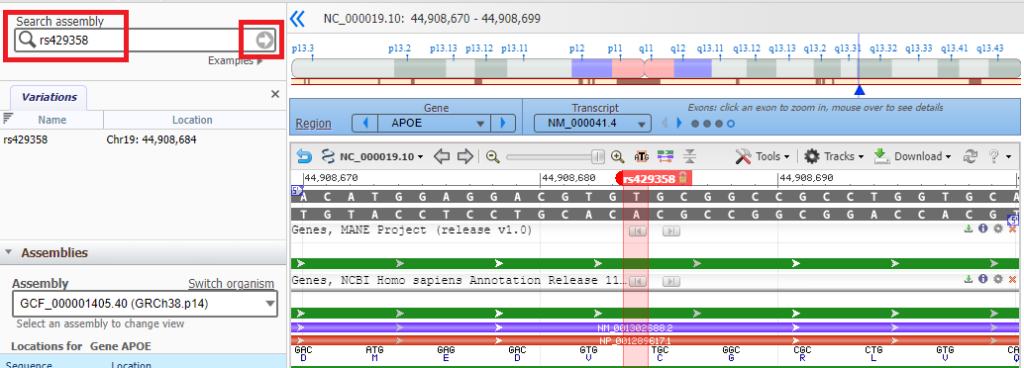
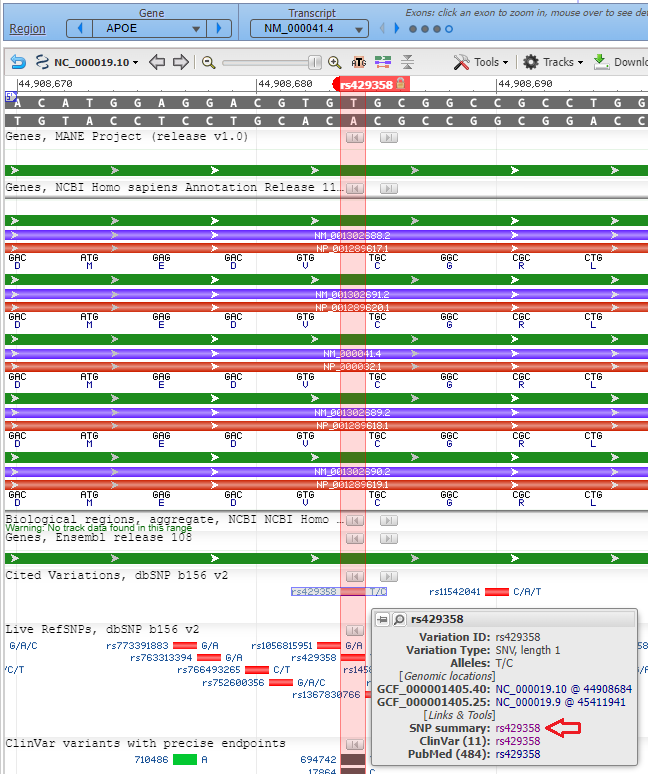
10. Click the Back button of your browser to return to the Genomic Data Viewer, then type (or copy and paste) rs429358 into the Search box as shown below and click the arrow in that box. Note that thymine (T) is present at this location on the top strand.
11. Click on the small red box labelled ‘rs429358‘ and a box will appear as shown at the bottom of the graphic below. Then click ‘rs429358‘ next to ‘SNP summary’, as shown at the red arrow.
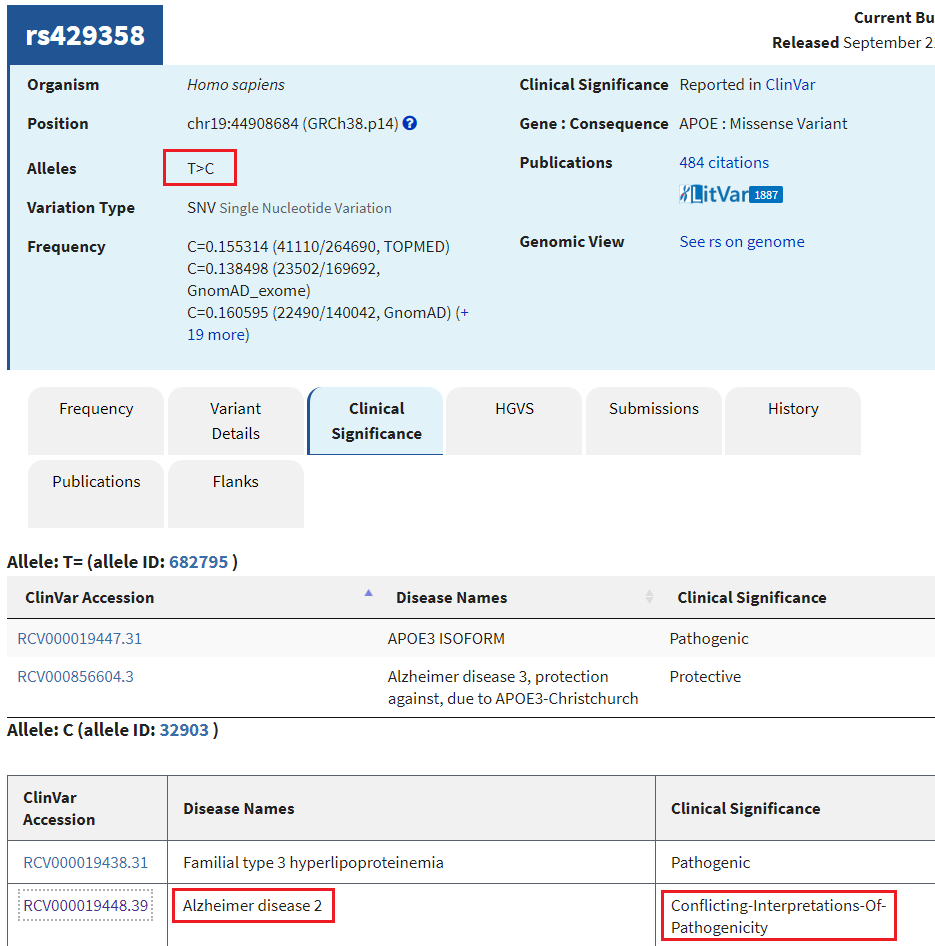
12. A new tab will appear on your browser showing this entry in the SNP database. T>C means that T can be changed to C at this location. Click on the Clinical Significance tab and note that the T > C mutation is associated with Alzheimer’s disease, although there are “conflicting interpretations of pathogenicity”.
Question: Should a genetic test be done to determine the likelihood that a person will come down with late-onset Alzheimer’s disease? Why or why not?
13. Click the Publications tab to see a list of publicaations specific to this particular SNP. Only a sampling is given here; the View All in Pubmed button gives access to the full list.
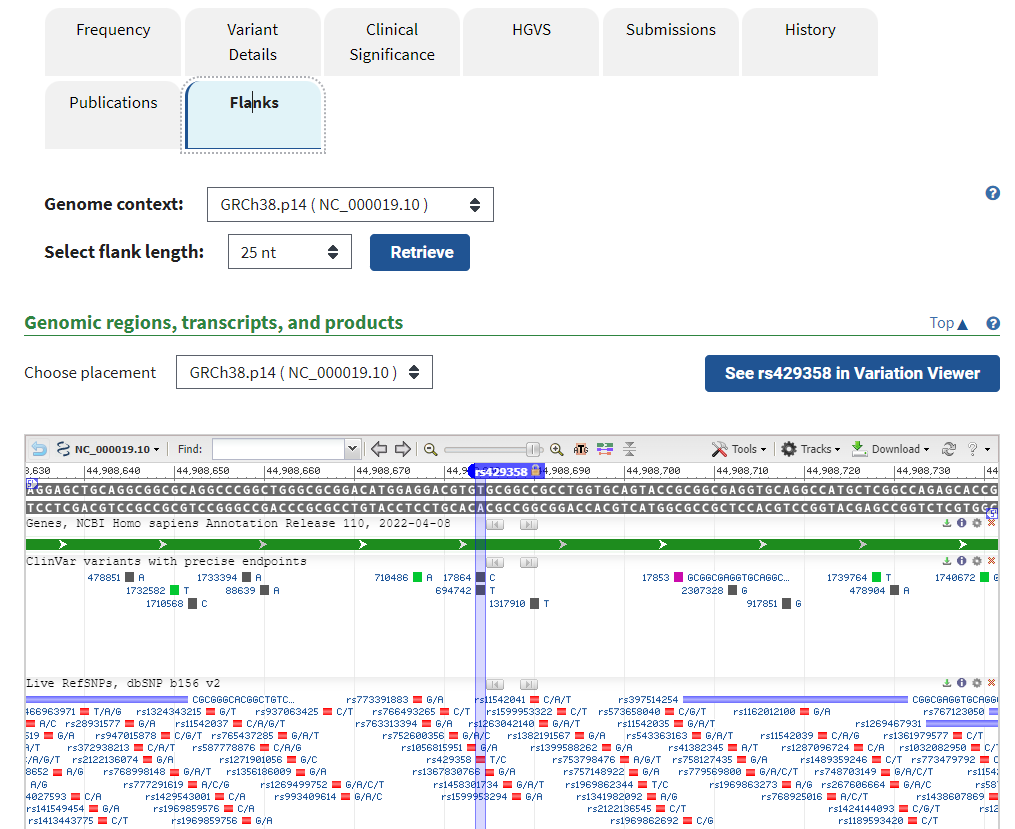
14. Click on the Flanks tab to see other SNPs identified for this portion of the APOE gene. Mouse over some of these to see if any have clinical significance (indicated by a ClinVar link).
Goal of Steps 15 – 24: Examine wild-type and mutant variations on the three-dimensional structure of the APOE protein.
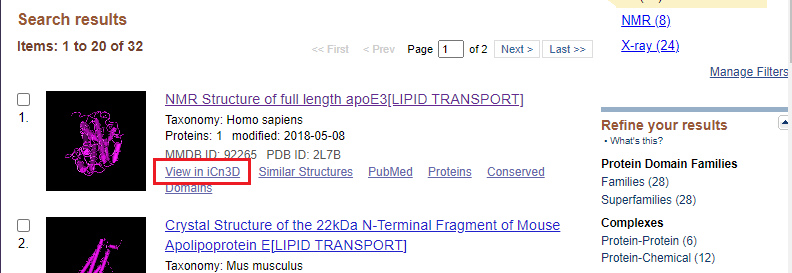
15. Click this link to open the NCBI Structure website, then type APOE into the search field and click the Search button.
16. The first link should be as shown below (if not, scroll down to ‘NMR Structure of full length apoE3[LIPID TRANSPORT]). Click on View in iCn3D for this link.
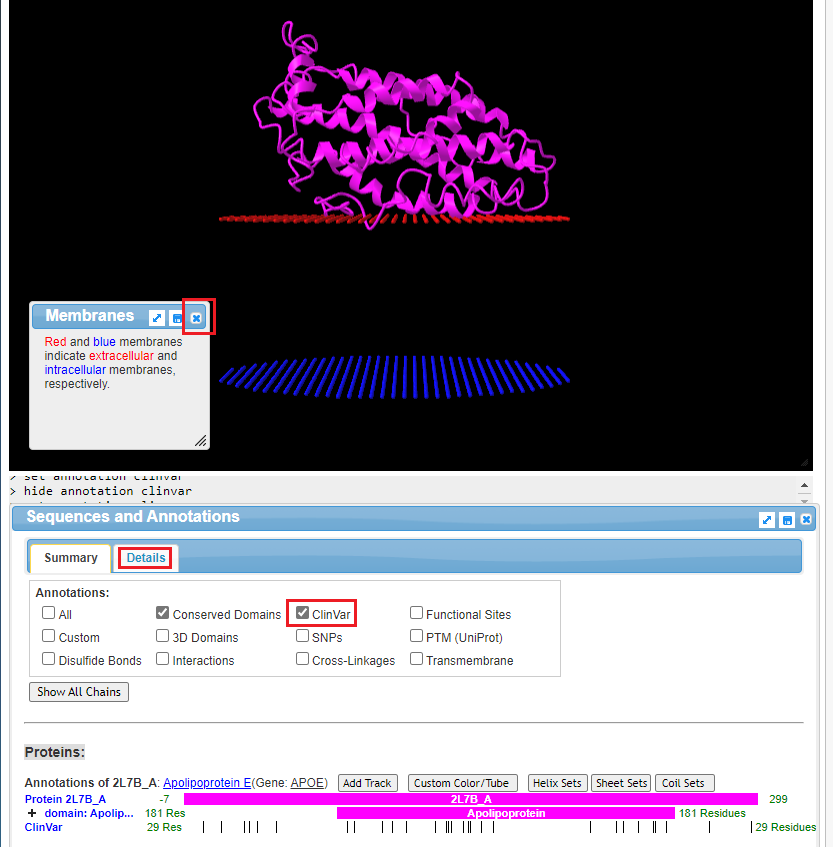
17. The structure of the protein is shown, along with portions of the cell membrane. Click on the ‘x’ to close the Membranes box. The structure can be rotated in three dimensions using your mouse cursor and wheel, and the entire structure can be moved using by right-clicking and dragging. Note: On a tablet or phone, use one finger to rotate the structure, and two fingers to move the entire structure. If the structure disappears when you attempt to manipulate it, hit the back button of your browser to go back to Step 16, and click on View in iCn3D again. You may have to experiment with finger movements, especially on a phone.
Click the ClinVar checkbox, then click the Details tab.
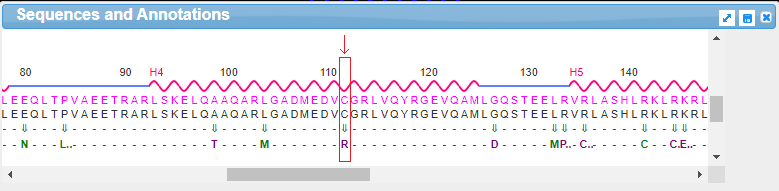
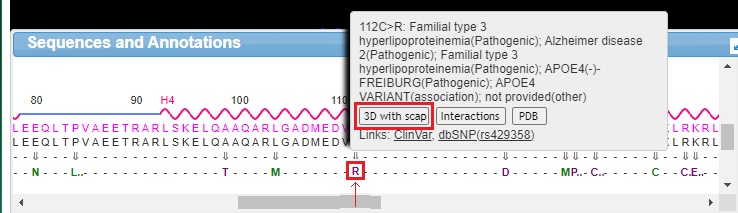
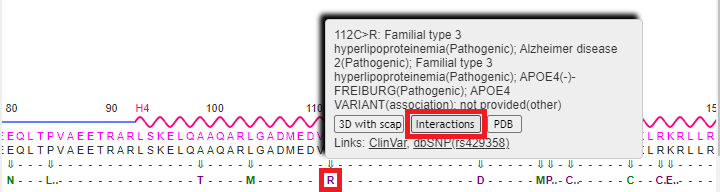
18. Drag the scrollbar at the bottom of the Sequences and Annotations window to scroll to location 112, shown below in the red box below. The ‘wild type’ amino acid at this location is cysteine (C), and the SNP variation (mutation) is arginine (R). Note: On a phone or tablet, use your finger to carefully drag the protein sequence.
Note: The codon associated with cysteine is TGT, and the codon associated with arginine is CGT – see Step 11 above. Don’t confuse the amino acid cysteine (C) with the nucleotide cytosine (also abbreviated C).
19. Mouse over the R without clicking, and a pop-up box will appear. Click the 3D with scap button in this box. Note: On a phone or tablet, delicately touch the R with your finger until the pop-up box appears.
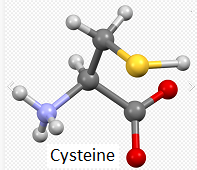
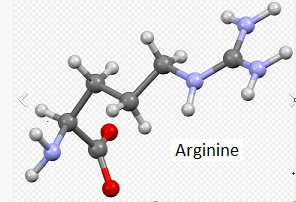
20. The three-dimensional structure that appears can be rotated by dragging with your mouse and enlarged using the wheel on your mouse. Move the model until the multicolored amino acid at location 112 is clearly visible, as shown in the screen shot below. Press the A key of your keyboard (or use the View menu on a tablet or phone*) to toggle back and forth between cysteine and arginine (see images below). As you rotate the image, note that peptide bonds link cysteine with adjacent amino acids in the protein.
Note: For context, unbound cysteine and arginine amino acids are also shown, with the colors the same as in the 3D image (see color code in 3D image).
*The View menu may have a different appearance on a phone – it is accessed via the small box with three lines at upper left.
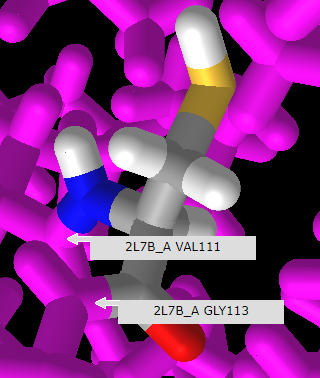
21. Rotate the model and mouse over the amino acids immediately adjoining cysteine or arginine to reveal that they are valine (V) at position 111 and glycine (G) at position 115, as shown below. Compare this with the protein sequence shown in step 18 above. Click here to see a video of this procedure, with arginine at the SNP position rather than cysteine.
Note: When forming peptide bonds, the amino (nitrogen containing) group of one amino acid is linked to the carboxyl (oxygen-containing) group of an adjacent amino acid. In the process, a water molecule is given off, so that is the reason only one oxygen is shown in the 3D model, whereas two are shown in the unbound cysteine and arginine ball-and-stick models.
22. Press the back button of your browser to repeat steps 16-20 above, only this time click on the Interactions button on the pop-up menu (it may not work properly unless you repeat these steps).
23. Press the A key of your keyboard (or use the View menu on a tablet or phone) to toggle back and forth between cysteine and arginine. The amino acids will appear with dotted lines showing their complex interactions with other amino acids in the protein. Drag with your mouse to rotate the 3D model and observe the interactions.
Questions:
1. How might these interactions determine the shape and function of the protein?
2. How might this be related to a possible link between SNP variations and late-onset Alzheimer’s disease?
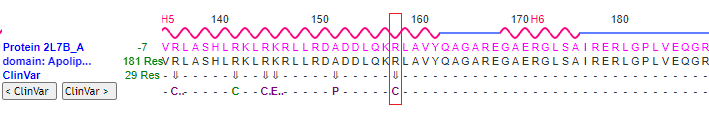
24. Use the above procedure (steps 16-23) to visualize the mutation associated with SNP rs7412, at position 158 on the protein. Use the 3D model to verify that the wild type has arginine at this position, and that the mutation results in arginine being replaced by cysteine being at that position (R > C). Also verify that the arginine or cysteine is flanked by lysine (K) at position 157, and leucine (L)at position 159. You will need to rotate the protein carefully to see these locations on the model.
Question:
1. If the mutation is arginine (R) to cysteine (C), which APOE allele would result from the mutation? Hint: Look at the table in Step 1, and remember that the codon for cysteine is TGC, and the codon for arginine is CGC. Again, don’t confuse the amino acid cysteine (C) with the nucleotide cytosine (also abbreviated C).
2. Which disease conditions are associated with the answer to the above question?
25. Click here to access a pioneering research paper that described the first use of base editing to correct SNP mutations. One goal of this research was to correct SNP mutations in mouse cells that had been transformed by replacing the mouse APOE gene with the human APOE4 allele. Specifically, the goal was the convert the APOE4 allele into the APOE3 allele. The relevant paragraph is reproduced below, located just above Table 1 in the linked paper. Note: The lead author is a researcher in the lab of Dr. David Liu. Click this link to view a YouTube video of a presentation by Dr. Liu describing base editing, prime editing, and other advances in this field.
“Finally, we tested the potential of base editing to correct two disease-relevant mutations in mammalian cells. The apolipoprotein E gene variant APOE4 encodes two Arg residues at amino acid positions 112 and 158, and is the largest and most common genetic risk factor for late-onset Alzheimer’s disease.23 ApoE variants with Cys residues at these positions, including APOE2 (Cys112/Cys158), APOE3 (Cys112/Arg158), and APOE3r (Arg112/Cys158) have been shown or are presumed24 to confer lower Alzheimer’s disease risk than APOE4. We attempted to convert APOE4 into APOE3r in immortalized mouse astrocytes in which the endogenous APOE gene was replaced by human APOE4. We delivered into these astrocytes by nucleofection DNA encoding BE3 and an appropriate sgRNA placing the target C at position 5 relative to a downstream PAM. After two days, we isolated nucleofected cells and measured editing efficiency by HTS of genomic DNA. We observed conversion of Arg158 to Cys158 in 58-75% of total DNA sequencing reads (Table 1a and Extended Data Fig. 8a). We also observed 36-50% editing of total DNA at the third position of codon 158 and 38-55% editing of total DNA at the first position of Leu159, as expected since all three of these Cs are within the base editing window. Neither of the other two C→T conversions, however, alters the amino acid sequence of the ApoE3r protein since both TGC and TGT encode Cys (all C→T changes at the third position of a codon are silent), and both CTG and TTG encode Leu.” Komer et al. 2016
Questions:
1. Based on these results, if you were in charge, would you ok the use of this treatment for use in human patients? Why or why not?
2. Consider this 2023 article on the safety of CRISPR editing in embryos. Do these concerns also apply to the potential use of base editing for Alheimer’s disease?