Background on the most common CF mutation (delta F508).
Goal of Steps 1-7: Use the Genome Data Viewer and SNP database to examine sequences associated with this mutation.
Goal of Steps 8-15: Use the NCBI Structure tool to locate the most common mutation on the three-dimensional structure of the CFTR protein.
Goal of Steps 16-17: Examine two additional CFTR mutations and learn about prime editing, a potential tool to correct these and other pathogenic mutations.
Return to Cases A, B, and C
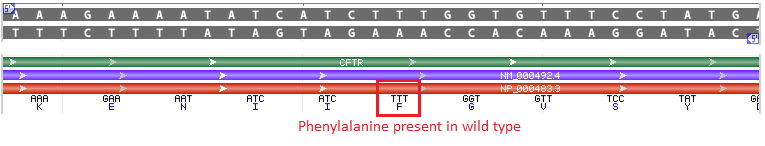
Background: CFTR is a protein responsible for the transport of chloride ions out of cells. A portion of the normal (wild type) CFTR protein is shown below in orange, with phenylalanine (F) present at position 508. TTT is the codon associated with phenylalanine. Gray lines and green lines represent the DNA sequence, whereas purple represents mRNA.
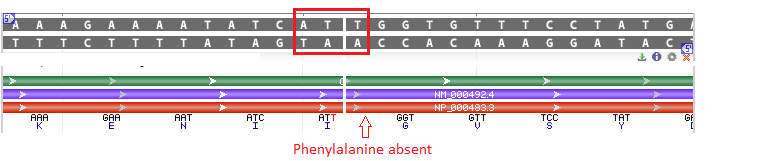
Cystic fibrosis is caused by mutations in the gene that codes for the CFTR protein. The most common CF mutation is a deletion of 3 base pairs in the CFTR gene at the location shown below.
The effect of this deletion is to convert an ATCTTT sequence (coding for isoleucine and phenylalanine) into an ATT sequence (coding for isoleucine). Note that both ATC and ATT code for isoleucine. The net effect of the deletion is to eliminate phenylalanine at position 508 of the CFTR protein, causing loss of function.
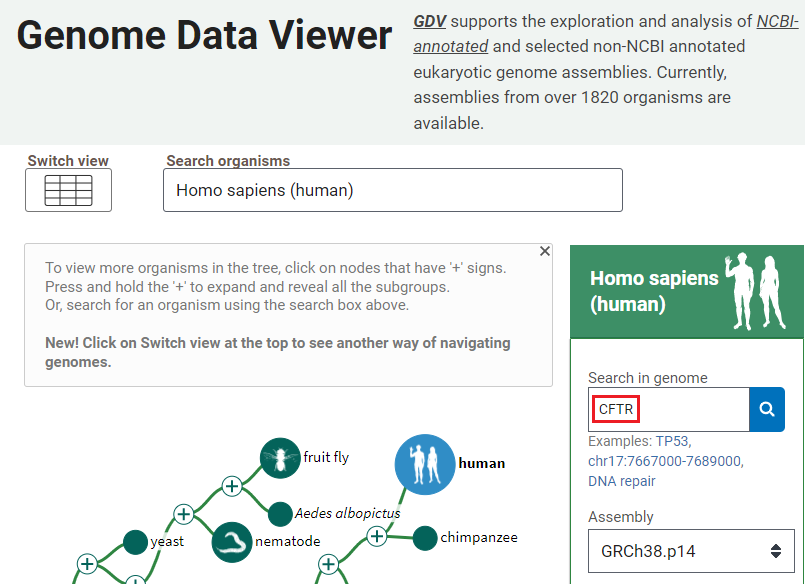
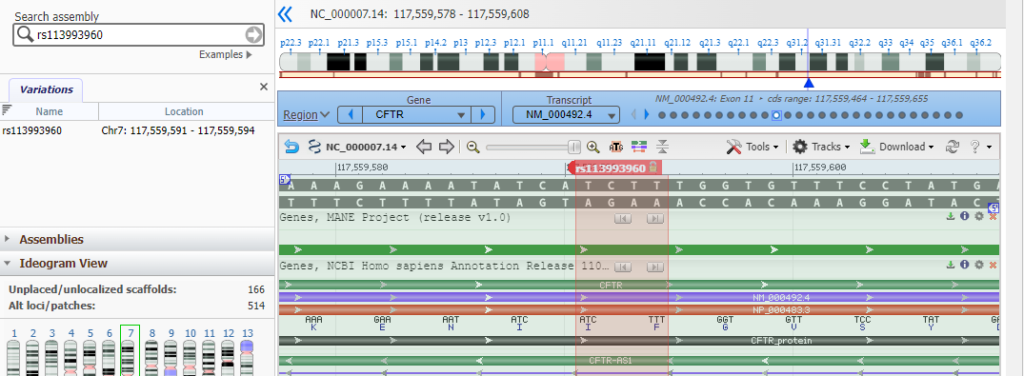
1. Open the Genome Data Viewer and enter CFTR into the search box, as shown below, and click the magnifying glass icon so search for this SNP. If you copy and paste, use control-V or command -V to paste.
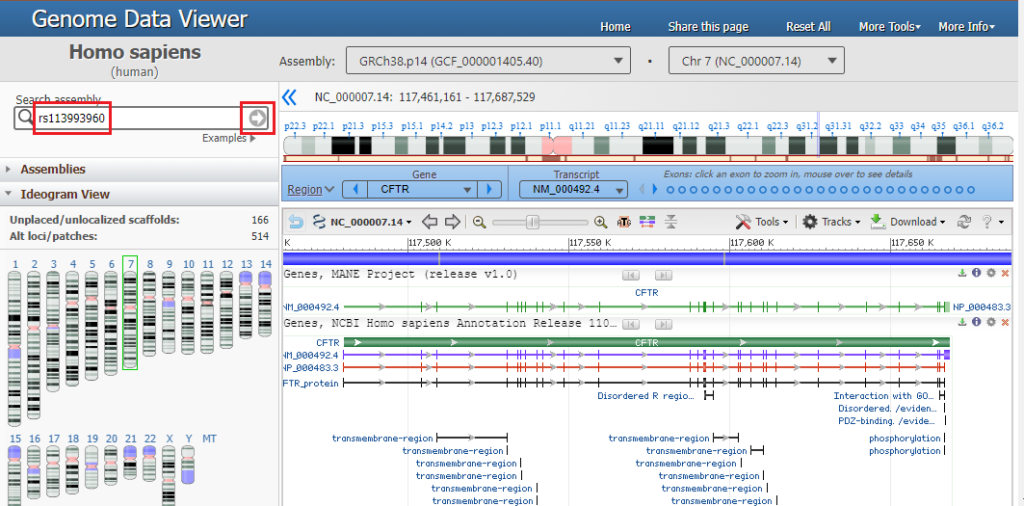
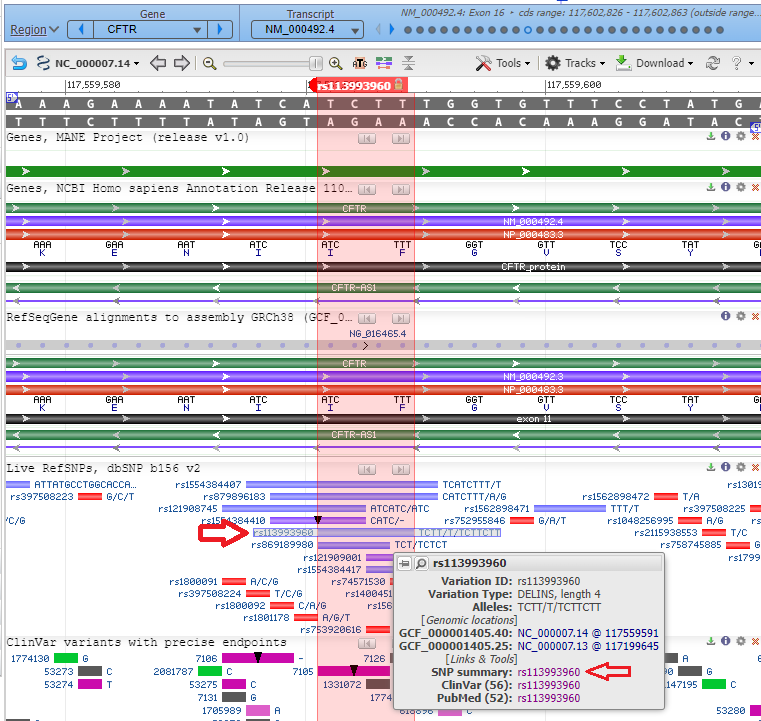
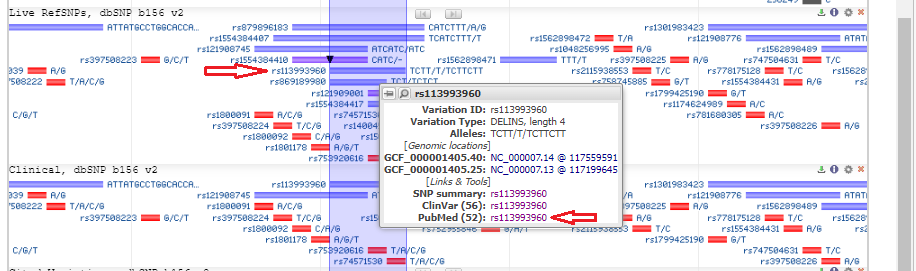
2. The CFTR gene appears in condensed format, with short vertical lines indicating the locations of the 27 exons. Note that the gene is located on chromosome 7. Type (or copy and paste) rs113993960 into the ‘search assembly’ box, then click the arrow in this box (first image below). This reference number is associated with the location of the mutation described above. A magnified region of the CFTR gene appears, at the location associated with this reference number (second image below).
3. Scroll down to the bar labelled rs113993960, hold your mouse over the bar, and click on the SNP summary link that appears in the pop-up window.
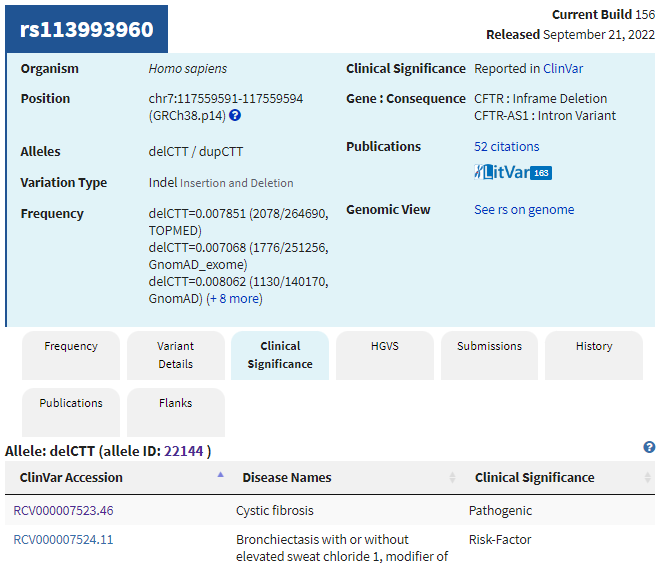
4. The summary appears in the dSNP database. Note that alleles are delCTT/dupCTT, meaning that the CTT triplet can either be deleted (as discussed above) or duplicated. Click the Clinical Significance tab to see a list of conditions associated with the delCTT allele.
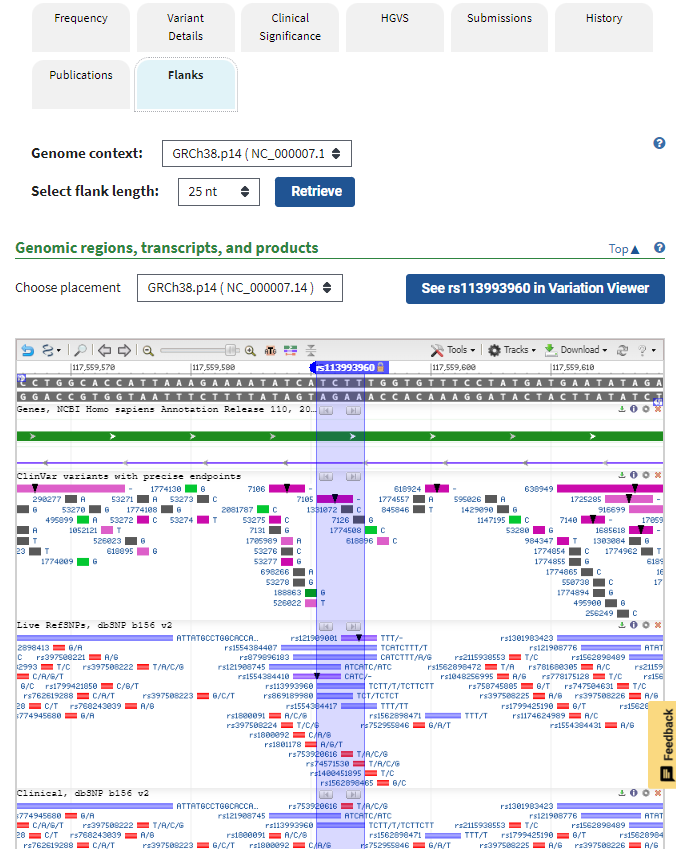
5. Click the Flanks tab and scroll down to see a display similar to a scaled-down version of the Genome Data Viewer. The link in the blue box allows you to see the same mutation in the Variation Viewer.
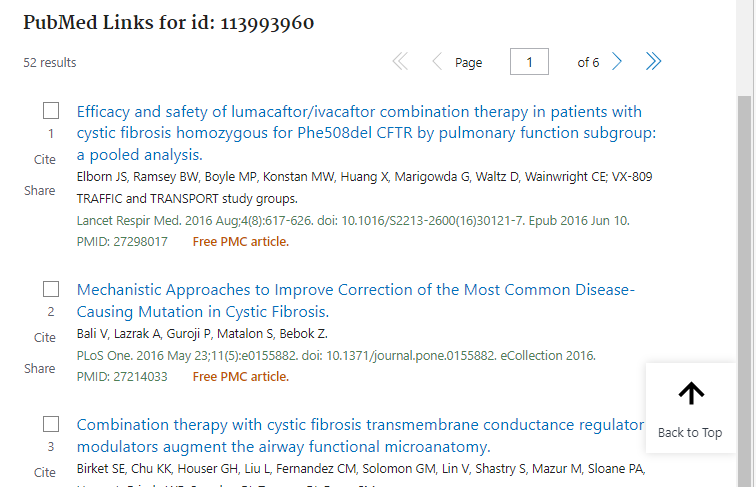
6. Hover your mouse over the rs113993960 bar until the pop-up box appears, then select the PubMed link as shown below.
Note: There are many ways to navigate using NCBI tools, for example by entering the rs113993960 reference number directly into the dSNP database.
7. A list of publications appears related to cystic fibrosis. You can also access related publications using the Publications tab (see Step 5 above).
Goal of Steps 8-15: Use the NCBI Structure tool to locate the mutation site on the three-dimensional structure of the CFTR protein.
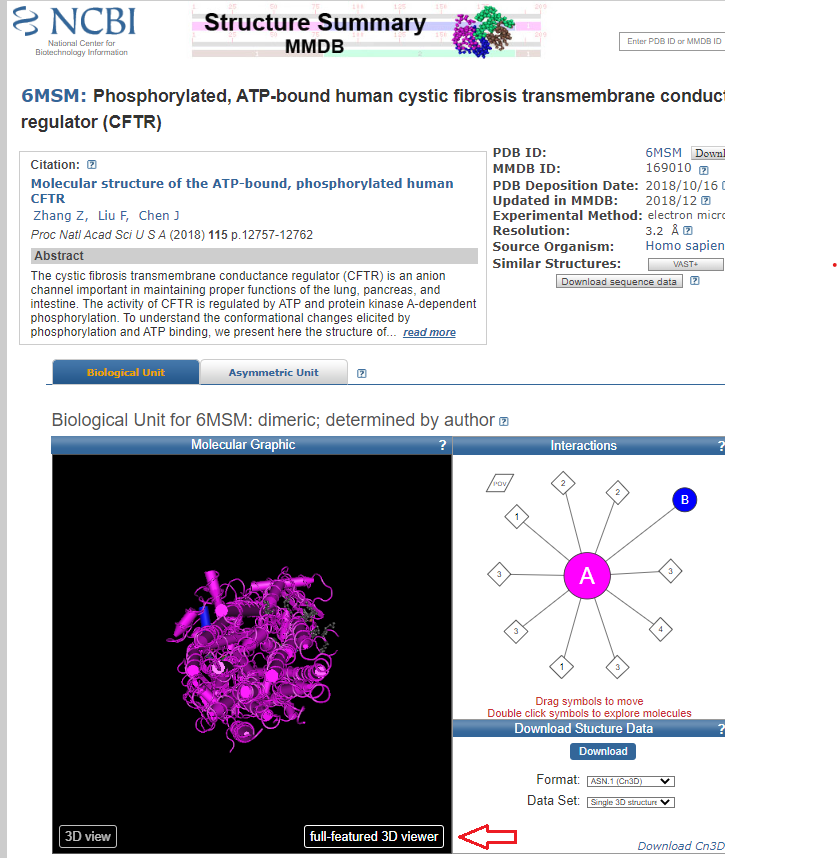
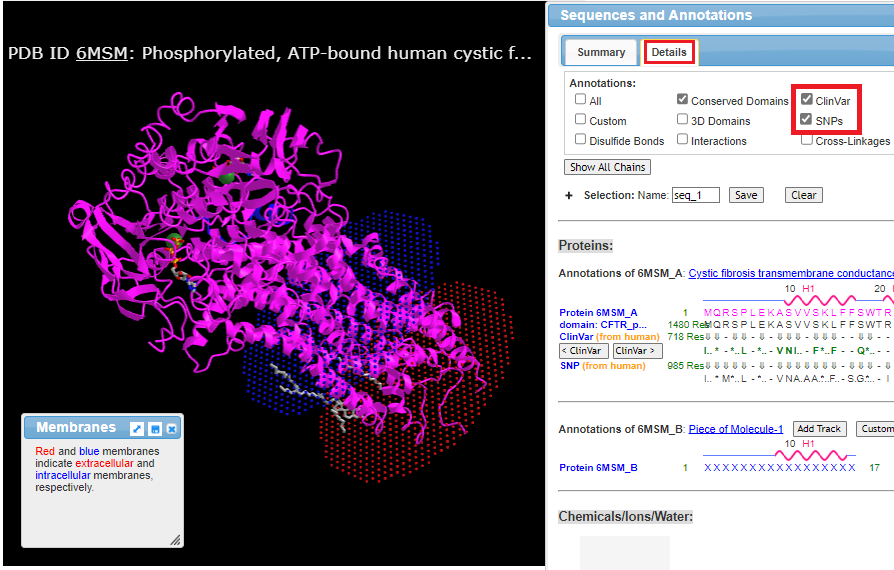
8. Click here to open the NCBI Structure home page, type 6MSM into the search field, and press the enter (or return) key. This is the reference code for the CFTR protein.
9. Click the full-featured 3D viewer button at the bottom of the protein image.
10. Rotate the structure with you mouse to see how it is embedded in the cell membrane (blue intracellular, red extracellular). The entire structure can be moved by holding down the right mouse button and dragging.
Check the ClinVar and SNPs boxes so that SNP locations and Clinvar annotations appear along with the protein sequence, then click the Details tab. Note that the Sequences and Annotations window is moveable and will first appear either at the bottom or the side of your browser window, depending on its configuration.
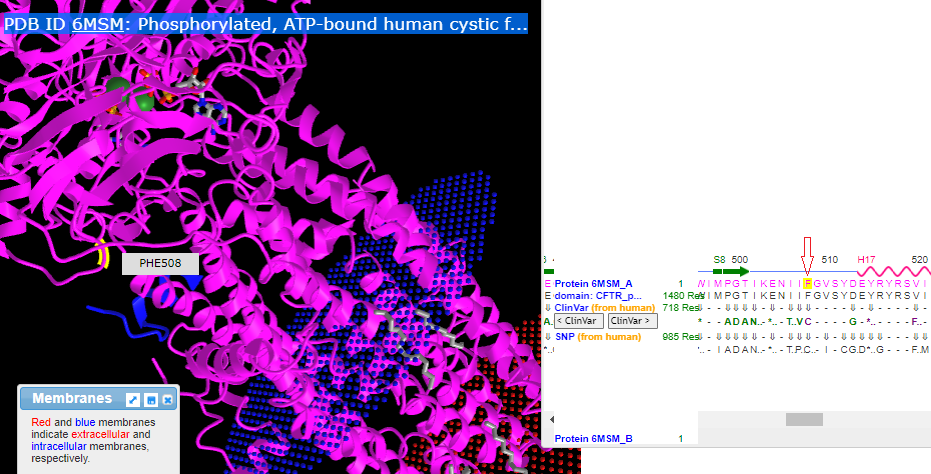
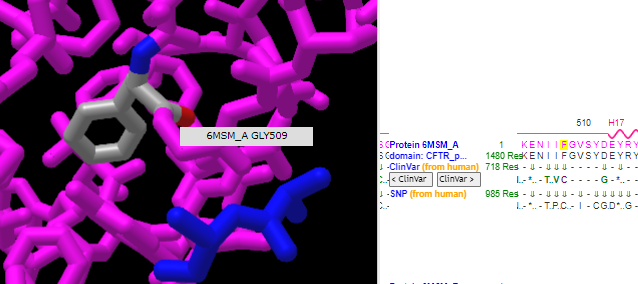
11. Use the dragbar (or your finger if using a phone) to slide the sequence to position 508, and carefully drag over (or tap) the F to highlight it as shown below, in yellow. Hover your mouse over the yellow region on the 3D structure, and a box will pop up verifying that this is phenylalanine at location 508. Note that F and PHE are both abbreviations for phenylalanine.
12. Click here for a video of the structure being rotated, showing the amino acids at positions 506-509 (isoleucine, isoleucine, phenylalanine, glycine). ATP (multicolored) and magnesium (green ball) are also shown in the video.
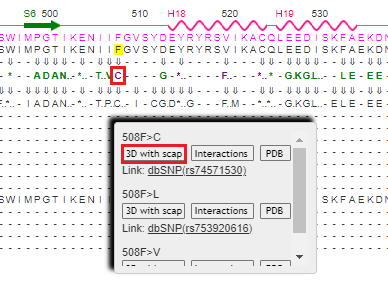
13. This particular model does not show the effect of phenylalanine (F) being eliminated from position 508, the most common CF mutation. There are many other mutations that occur in the CFTR protein, however, some of which are benign and some of which are pathogenic. One of these is a SNP mutation where phenylalanine is replaced by cysteine (C) at position 508. To see this in 3D, hover your mouse over the C and click 3D with scap.
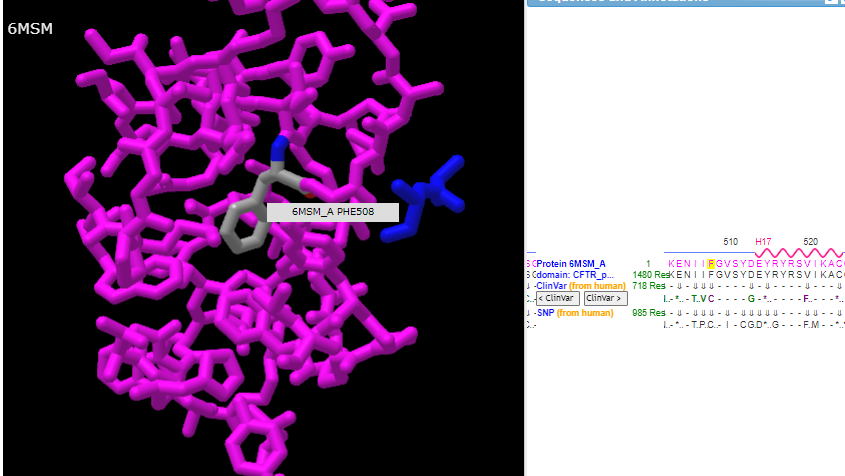
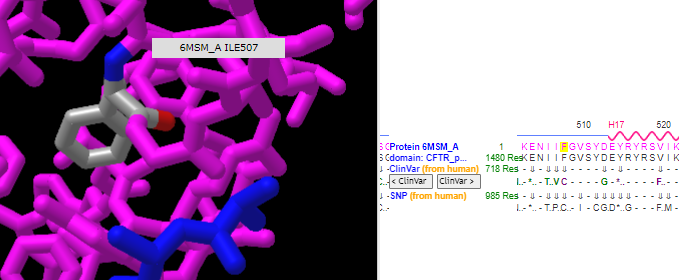
14. Hover your mouse over the phenylalanine at position 508 and verify again that isoleucine is at position 507 and glycine is at position 509, as shown in the three graphics below.
15. To alternate between phenylalanine to cysteine at position 508, use the View menu and select Alternate (key A) or press the A key of your computer keyboard. Click this link to view a video of this process. Note that on a phone or tablet, the menu is activated by a button (with three lines) in the upper-left corner of the screen.
Goal of Steps 16-17: Examine two additional CF mutations and learn about prime editing, a potential tool to correct these and other pathogenic mutations.
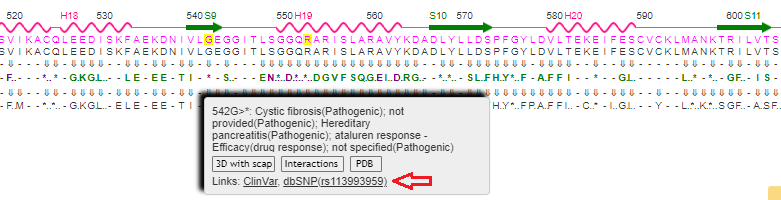
16. Scan the protein sequence to find glycine (G) at location 542 and hover over the ClinVar line. Here is a direct link to the Variant Details for rs113993959. Note that there are two alleles associated with this mutation, G>A and G>T, resulting in a change from glycine [GGA] to argine [AGA], or glycine [GGA] to TGA, a stop codon.
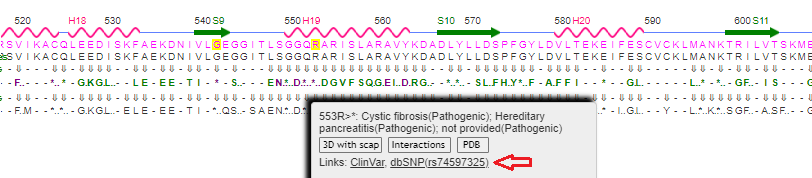
17. Scan the protein sequence again to find arginine (R) at location 553 and hover over the ClinVar line. Here is a direct link to the Variant Details for rs74597325. Note that there are two alleles associated with this mutation, C>G and C>T, resulting in a change from arginine [CGA] to glycine [GGA], or arginine [CGA] to TGA, a stop codon.
Delta F508 is the most common CF mutation, occurring in 70% of cases, but many other mutations occur such as G542X and R553X, described above. These mutations could potentially be corrected via prime editing, a modification of CRISPR genome editing.
Click here for an explanation of prime editing that includes the correction of G542X and R553X mutations and other research applications.
Click here to access the pioneering research paper on prime editing.
Note: The lead author is a researcher in the lab of Dr. David Liu. Click this link to view a YouTube video of a presentation by Dr. Liu describing base editing, prime editing, and other advances in this field.
Questions:
1. Hover your mouse cursor over other mutations of the CFTR protein sequence. How many can you find that are associated with cystic fibrosis or related pathogenic conditions? How many mutations are benign?
2. Why is prime editing considered to be safer than the original, CRISPR Cas9 technique for genome editing?